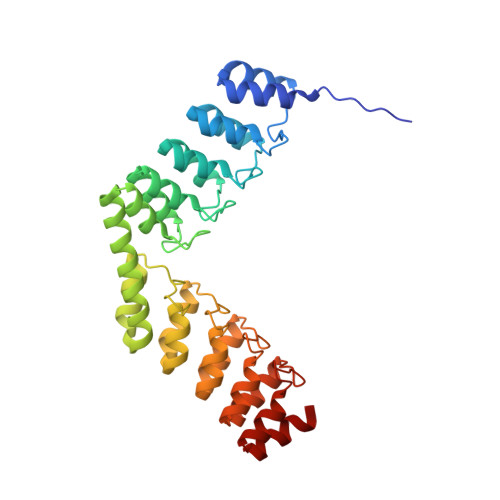
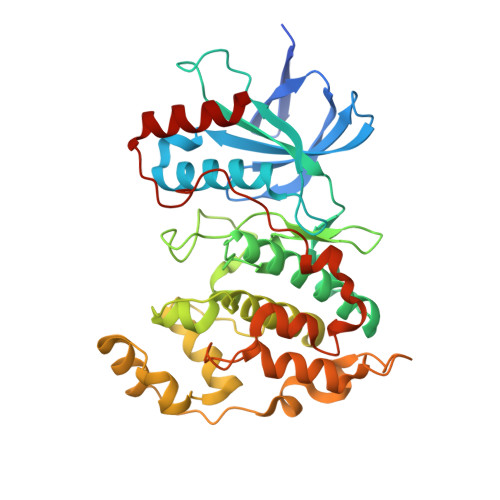
Structural Basis for the Selective Inhibition of c-Jun N-Terminal Kinase 1 Determined by Rigid DARPin-DARPin Fusions.
Wu, Y., Honegger, A., Batyuk, A., Mittl, P.R.E., Pluckthun, A.(2018) J Mol Biol 430: 2128-2138
- PubMed: 29126898
- DOI: https://doi.org/10.1016/j.jmb.2017.10.032
- Primary Citation of Related Structures:
5LW1 - PubMed Abstract:
To untangle the complex signaling of the c-Jun N-terminal kinase (JNK) isoforms, we need tools that can selectively detect and inhibit individual isoforms. Because of the high similarity between JNK1, JNK2 and JNK3, it is very difficult to generate small-molecule inhibitors with this discriminatory power. Thus, we have recently selected protein binders from the designed ankyrin repeat protein (DARPin) library which were indeed isoform-specific inhibitors of JNK1 with low nanomolar potency. Here we provide the structural basis for their isotype discrimination and their inhibitory action. All our previous attempts to generate crystal structures of complexes had failed. We have now made use of a technology we recently developed which consists of rigid fusion of an additional special DARPin, which acts as a crystallization enhancer. This can be rigidly fused with different geometries, thereby generating a range of alternative crystal packings. The structures reveal the molecular basis for isoform specificity of the DARPins and their ability to prevent JNK activation and may thus form the basis of further investigation of the JNK family as well as novel approaches to drug design.
Organizational Affiliation:
Department of Biochemistry, University of Zürich, Winterthurerstrasse 190, CH-8057 Zürich, Switzerland.