Molecular insights into the mechanism of 4-hydroxyphenylpyruvate dioxygenase inhibition: enzyme kinetics, X-ray crystallography and computational simulations.
Lin, H.Y., Yang, J.F., Wang, D.W., Hao, G.F., Dong, J.Q., Wang, Y.X., Yang, W.C., Wu, J.W., Zhan, C.G., Yang, G.F.(2019) FEBS J 286: 975-990
- PubMed: 30632699
- DOI: https://doi.org/10.1111/febs.14747
- Primary Citation of Related Structures:
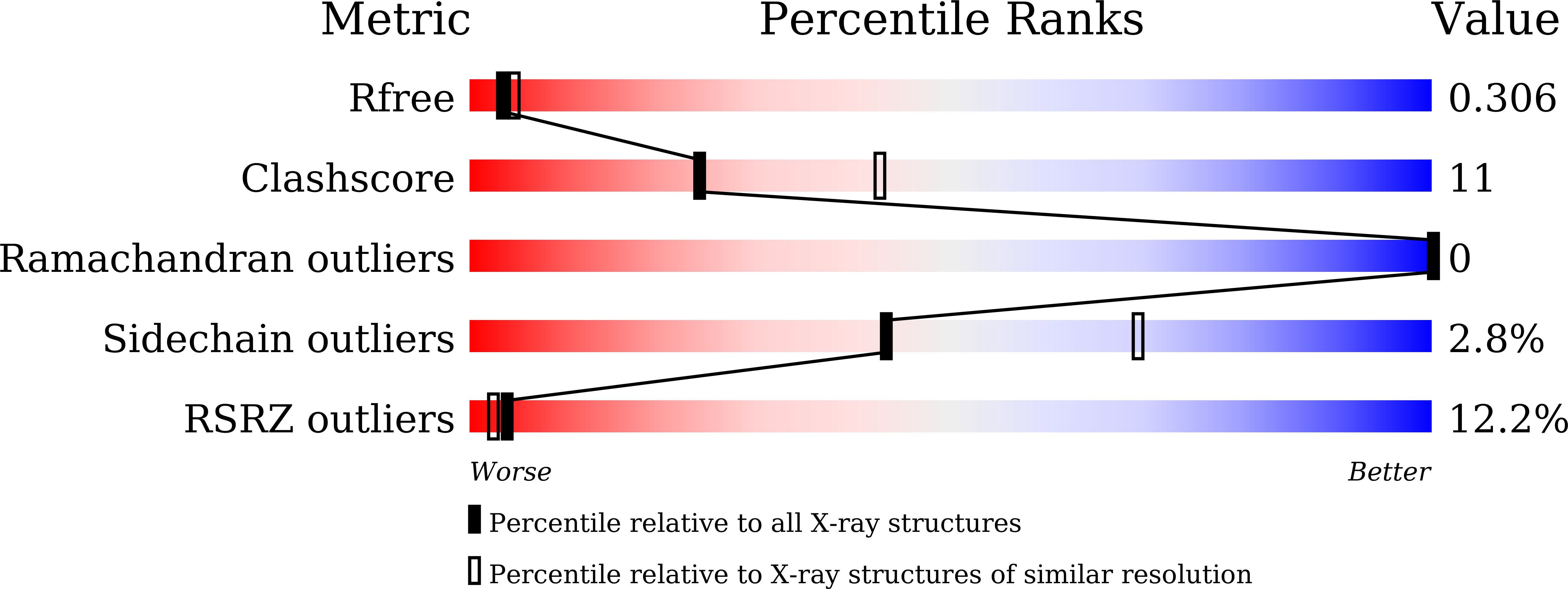
5YWG, 6ISD, 6J63 - PubMed Abstract:
Slow-binding inhibitors with long residence time on the target often display superior efficacy in vivo. Rationally designing inhibitors with low off-target rates is restricted by a limited understanding of the structural basis of slow-binding inhibition kinetics in enzyme-drug interactions. 4-Hydroxyphenylpyruvate dioxygenase (HPPD) is an important target for drug and herbicide development. Although the time-dependent behavior of HPPD inhibitors has been studied for decades, its structural basis and mechanism remain unclear. Herein, we report a detailed experimental and computational study that explores structures for illustrating the slow-binding inhibition kinetics of HPPD. We observed the conformational change of Phe428 at the C-terminal α-helix in the inhibitor-bound structures and further identified that the inhibition kinetics of drugs are related to steric hindrance of Phe428. These detailed structural and mechanistic insights illustrate that steric hindrance is highly associated with the time-dependent behavior of HPPD inhibitors. These findings may enable rational design of new potent HPPD-targeted drugs or herbicides with longer target residence time and improved properties. DATABASE: Structure data are available in the PDB under the accession numbers 5CTO (released), 5DHW (released), and 5YWG (released).
Organizational Affiliation:
Key Laboratory of Pesticide & Chemical Biology of Ministry of Education, International Joint Research Center for Intelligent Biosensor Technology and Health, Chemical Biology Center, College of Chemistry, Central China Normal University, Wuhan, China.