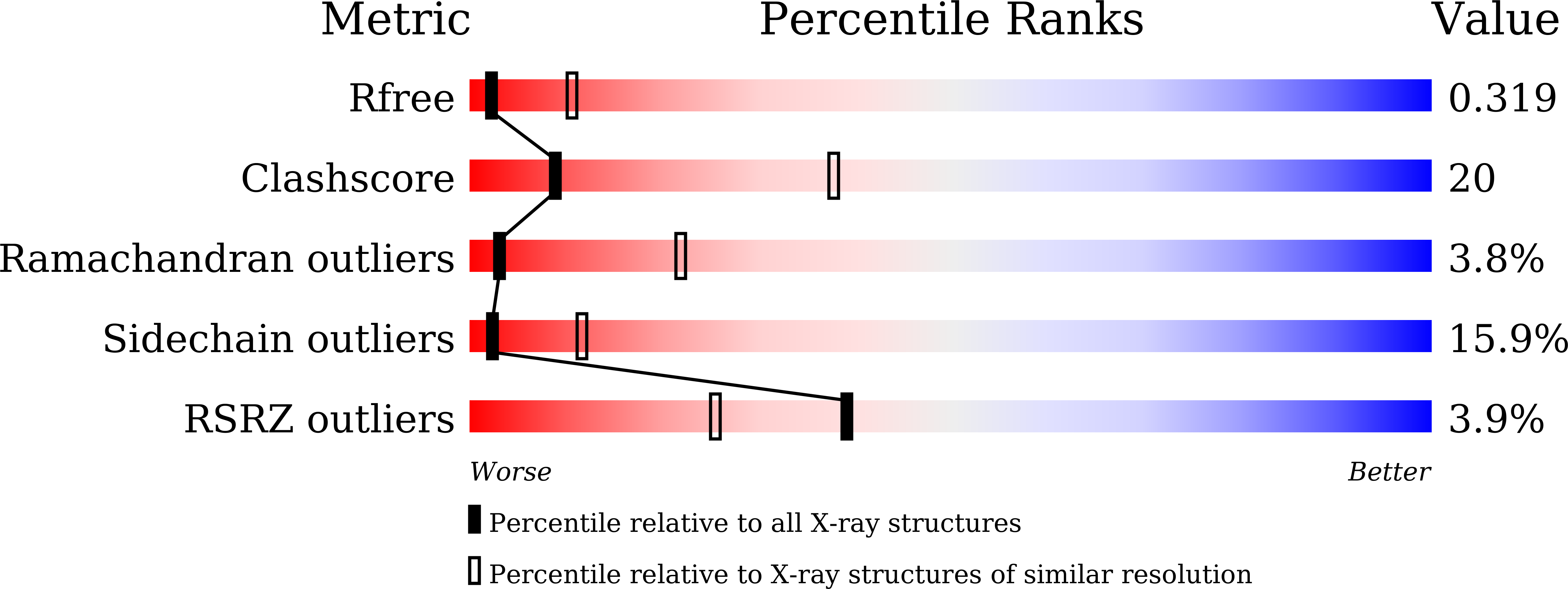
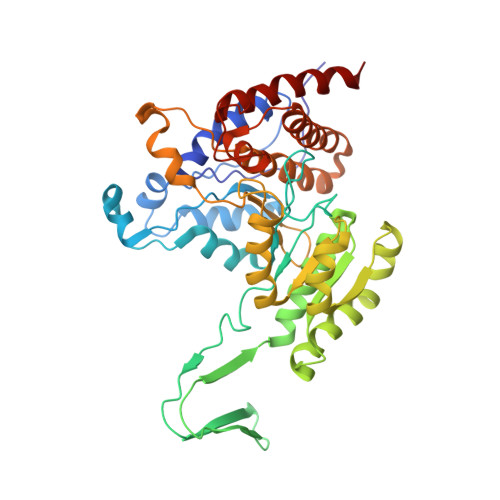
Biochemical characterization of a novel Trypanosoma brucei glycosomal isocitrate dehydrogenase with dual coenzyme specificity (NADP+/NAD+)
Wang, X., Inaoka, D.K., Shiba, T., Balogun, E.O., Ziebart, N., Allman, S., Watanabe, Y., Nozaki, T., Boshart, M., Bringaud, F., Harada, S., Kita, K.To be published.