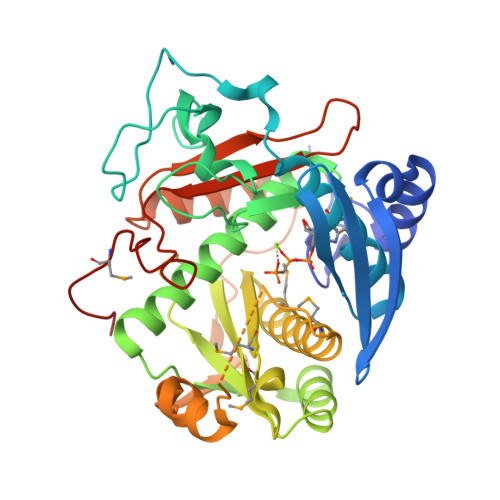
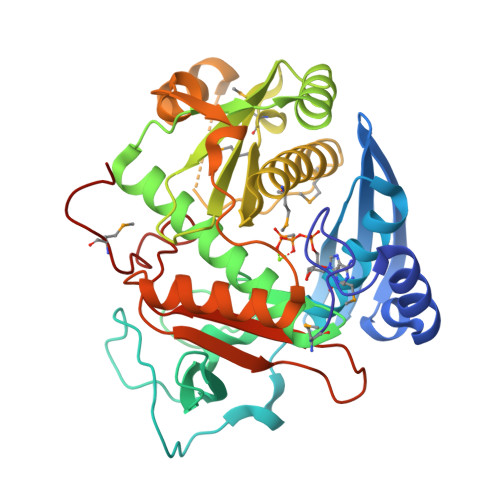
Enzymatic reconstitution of ribosomal peptide backbone thioamidation.
Mahanta, N., Liu, A., Dong, S., Nair, S.K., Mitchell, D.A.(2018) Proc Natl Acad Sci U S A 115: 3030-3035
- PubMed: 29507203
- DOI: https://doi.org/10.1073/pnas.1722324115
- Primary Citation of Related Structures:
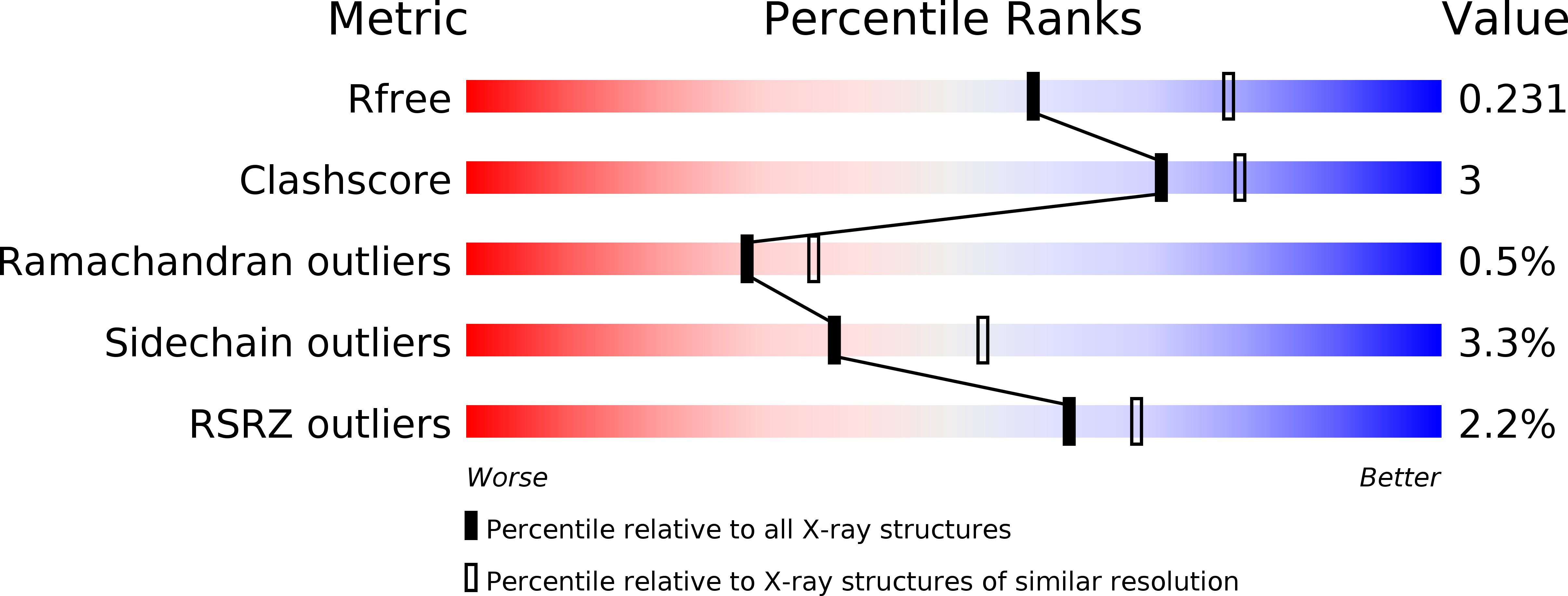
6CI7, 6CIB - PubMed Abstract:
Methyl-coenzyme M reductase (MCR) is an essential enzyme found strictly in methanogenic and methanotrophic archaea. MCR catalyzes a reversible reaction involved in the production and consumption of the potent greenhouse gas methane. The α-subunit of this enzyme (McrA) contains several unusual posttranslational modifications, including the only known naturally occurring example of protein thioamidation. We have recently demonstrated by genetic deletion and mass spectrometry that the tfuA and ycaO genes of Methanosarcina acetivorans are involved in thioamidation of Gly465 in the MCR active site. Modification to thioGly has been postulated to stabilize the active site structure of MCR. Herein, we report the in vitro reconstitution of ribosomal peptide thioamidation using heterologously expressed and purified YcaO and TfuA proteins from M. acetivorans Like other reported YcaO proteins, this reaction is ATP-dependent but requires an external sulfide source. We also reconstitute the thioamidation activity of two TfuA-independent YcaOs from the hyperthermophilic methanogenic archaea Methanopyrus kandleri and Methanocaldococcus jannaschii Using these proteins, we demonstrate the basis for substrate recognition and regioselectivity of thioamide formation based on extensive mutagenesis, biochemical, and binding studies. Finally, we report nucleotide-free and nucleotide-bound crystal structures for the YcaO proteins from M. kandleri Sequence and structure-guided mutagenesis with subsequent biochemical evaluation have allowed us to assign roles for residues involved in thioamidation and confirm that the reaction proceeds via backbone O -phosphorylation. These data assign a new biochemical reaction to the YcaO superfamily and paves the way for further characterization of additional peptide backbone posttranslational modifications.
Organizational Affiliation:
Department of Chemistry, University of Illinois, Urbana, IL 61801.