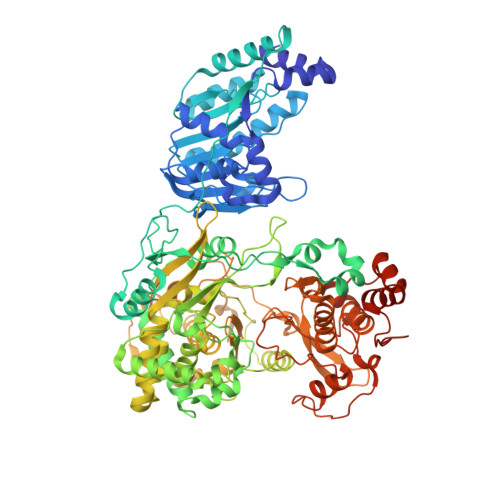
Supramolecular arrangement of the full-length Zika virus NS5.
Ferrero, D.S., Ruiz-Arroyo, V.M., Soler, N., Uson, I., Guarne, A., Verdaguer, N.(2019) PLoS Pathog 15: e1007656-e1007656
- PubMed: 30951555
- DOI: https://doi.org/10.1371/journal.ppat.1007656
- Primary Citation of Related Structures:
5M2X, 5M2Z, 6I7P - PubMed Abstract:
Zika virus (ZIKV), a member of the Flaviviridae family, has emerged as a major public health threat, since ZIKV infection has been connected to microcephaly and other neurological disorders. Flavivirus genome replication is driven by NS5, an RNA-dependent RNA polymerase (RdRP) that also contains a N-terminal methyltransferase domain essential for viral mRNA capping. Given its crucial roles, ZIKV NS5 has become an attractive antiviral target. Here, we have used integrated structural biology approaches to characterize the supramolecular arrangement of the full-length ZIKV NS5, highlighting the assembly and interfaces between NS5 monomers within a dimeric structure, as well as the dimer-dimer interactions to form higher order fibril-like structures. The relative orientation of each monomer within the dimer provides a model to explain the coordination between MTase and RdRP domains across neighboring NS5 molecules and mutational studies underscore the crucial role of the MTase residues Y25, K28 and K29 in NS5 dimerization. The basic residue K28 also participates in GTP binding and competition experiments indicate that NS5 dimerization is disrupted at high GTP concentrations. This competition represents a first glimpse at a molecular level explaining how dimerization might regulate the capping process.
Organizational Affiliation:
Structural Biology Unit, Institut de Biología Molecular de Barcelona CSIC, Barcelona, Spain.