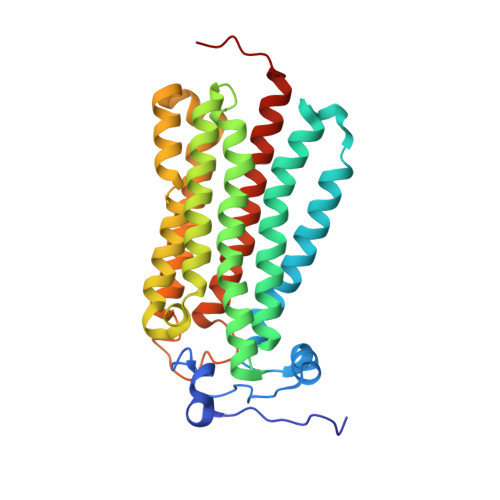
Human adiponectin receptor AdipoR1 assumes closed and open structures.
Tanabe, H., Fujii, Y., Okada-Iwabu, M., Iwabu, M., Kano, K., Kawana, H., Hato, M., Nakamura, Y., Terada, T., Kimura-Someya, T., Shirouzu, M., Kawano, Y., Yamamoto, M., Aoki, J., Yamauchi, T., Kadowaki, T., Yokoyama, S.(2020) Commun Biol 3: 446-446
- PubMed: 32796916
- DOI: https://doi.org/10.1038/s42003-020-01160-4
- Primary Citation of Related Structures:
6KRZ, 6KS0, 6KS1 - PubMed Abstract:
The human adiponectin receptors, AdipoR1 and AdipoR2, are key anti-diabetic molecules. We previously reported the crystal structures of human AdipoR1 and AdipoR2, revealing that their seven transmembrane helices form an internal closed cavity (the closed form). In this study, we determined the crystal structure of the D208A variant AdipoR1, which is fully active with respect to the major downstream signaling. Among the three molecules in the asymmetric unit, two assume the closed form, and the other adopts the open form with large openings in the internal cavity. Between the closed- and open-form structures, helices IV and V are tilted with their intracellular ends shifted by about 4 and 11 Å, respectively. Furthermore, we reanalyzed our previous wild-type AdipoR1 diffraction data, and determined a 44:56 mixture of the closed and open forms, respectively. Thus, we have clarified the closed-open interconversion of AdipoR1, which may be relevant to its functional mechanism(s).
Organizational Affiliation:
RIKEN Structural Biology Laboratory, 1-7-22 Suehiro-cho, Tsurumi-ku, Yokohama, 230-0045, Japan.