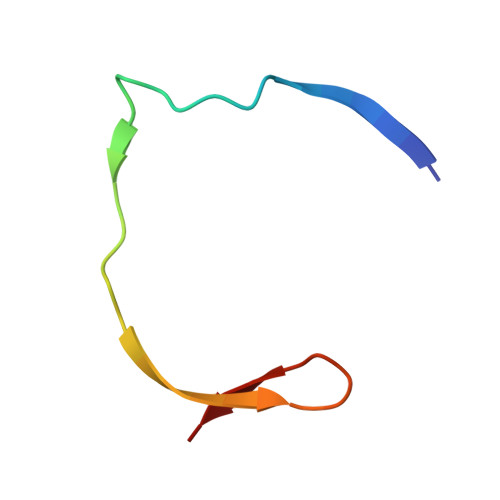
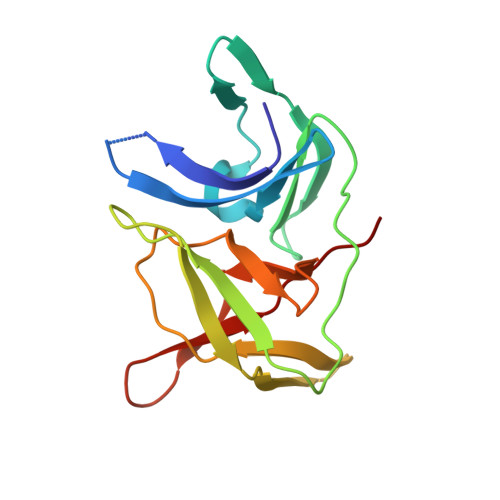
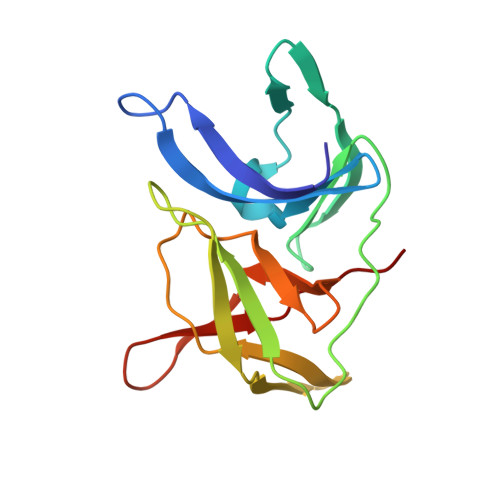
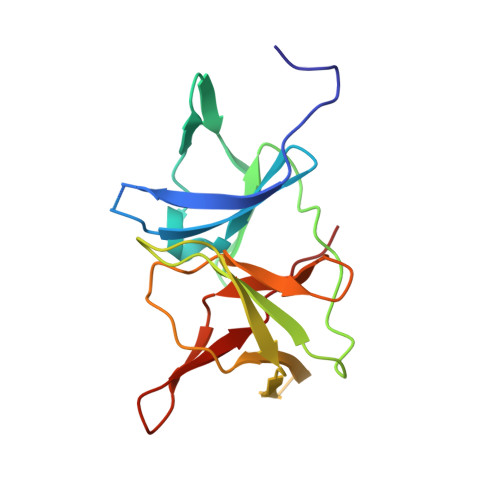
Identification and structural characterization of small molecule fragments targeting Zika virus NS2B-NS3 protease.
Quek, J.P., Liu, S., Zhang, Z., Li, Y., Ng, E.Y., Loh, Y.R., Hung, A.W., Luo, D., Kang, C.(2020) Antiviral Res 175: 104707-104707
- PubMed: 31953156
- DOI: https://doi.org/10.1016/j.antiviral.2020.104707
- Primary Citation of Related Structures:
6L4Z, 6L50 - PubMed Abstract:
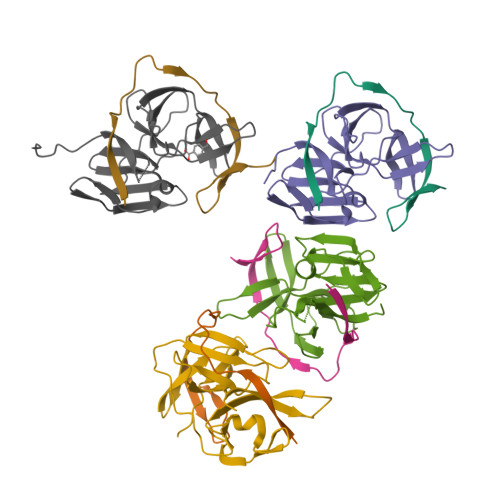
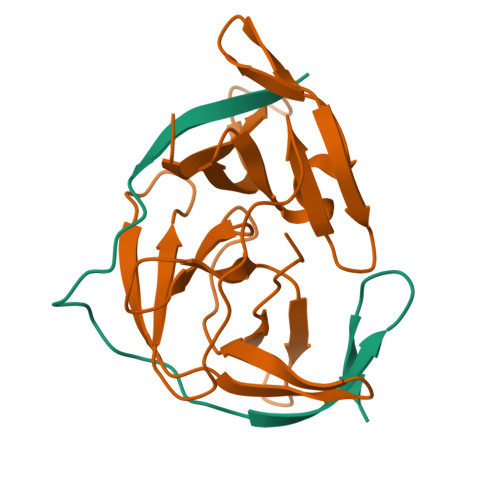
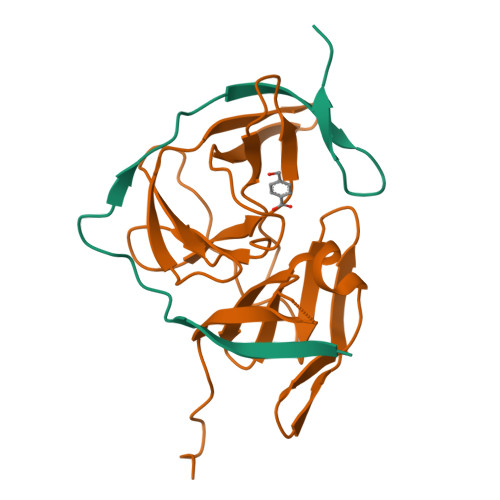
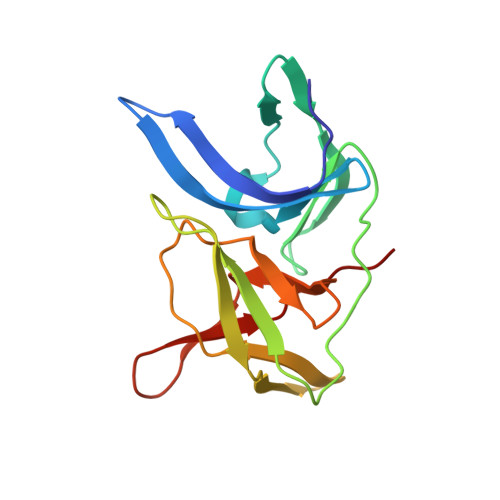
Zika virus (ZIKV) NS2B-NS3 protease is a validated antiviral target as it is essential for maturation of viral proteins. However, its negatively charged active site hinders the development of orthosteric small-molecule inhibitors. Fragment-based drug discovery (FBDD) is a powerful tool to generate novel chemical starting points against difficult drug targets. In this study, we scre ened a fragment compound library against the Zika protease using a primary thermal shift assay and identified twenty-two fragments which (bind to and) stabilize the protease. We then determined the X-ray crystal structures of two hits from different classes, all of which bind to the S1 pocket of the protease. We confirmed that these two fragments bind to the protease without inducing significant conformational changes using solution NMR spectroscopy. These fragment scaffolds serve as the starting point for subsequent lead compound development.
Organizational Affiliation:
Lee Kong Chian School of Medicine, Nanyang Technological University, EMB 03-07, 59 Nanyang Drive, Singapore, 636921, Singapore.