Discovery of a Novel Chemotype of Histone Lysine Methyltransferase EHMT1/2 (GLP/G9a) Inhibitors: Rational Design, Synthesis, Biological Evaluation, and Co-crystal Structure.
Milite, C., Feoli, A., Horton, J.R., Rescigno, D., Cipriano, A., Pisapia, V., Viviano, M., Pepe, G., Amendola, G., Novellino, E., Cosconati, S., Cheng, X., Castellano, S., Sbardella, G.(2019) J Med Chem 62: 2666-2689
- PubMed: 30753076
- DOI: https://doi.org/10.1021/acs.jmedchem.8b02008
- Primary Citation of Related Structures:
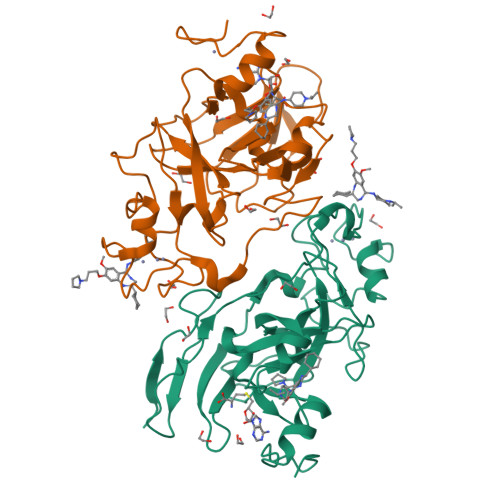
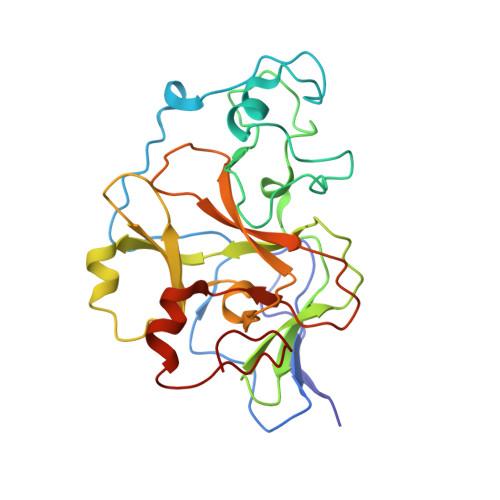
6MBO, 6MBP - PubMed Abstract:
Since the discovery of compound BIX01294 over 10 years ago, only a very limited number of nonquinazoline inhibitors of H3K9-specific methyltransferases G9a and G9a-like protein (GLP) have been reported. Herein, we report the identification of a novel chemotype for G9a/GLP inhibitors, based on the underinvestigated 2-alkyl-5-amino- and 2-aryl-5-amino-substituted 3 H-benzo[ e][1,4]diazepine scaffold. Our research efforts resulted in the identification 12a (EML741), which not only maintained the high in vitro and cellular potency of its quinazoline counterpart, but also displayed improved inhibitory potency against DNA methyltransferase 1, improved selectivity against other methyltransferases, low cell toxicity, and improved apparent permeability values in both parallel artificial membrane permeability assay (PAMPA) and blood-brain barrier-specific PAMPA, and therefore might potentially be a better candidate for animal studies. Finally, the co-crystal structure of GLP in complex with 12a provides the basis for the further development of benzodiazepine-based G9a/GLP inhibitors.
Organizational Affiliation:
Department of Molecular and Cellular Oncology , The University of Texas MD Anderson Cancer Center , Houston , Texas 77030 , United States.