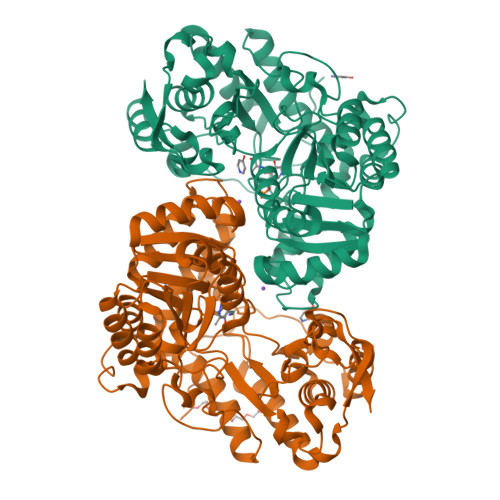
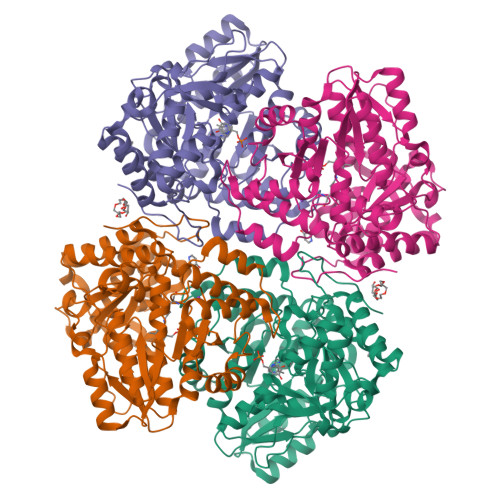
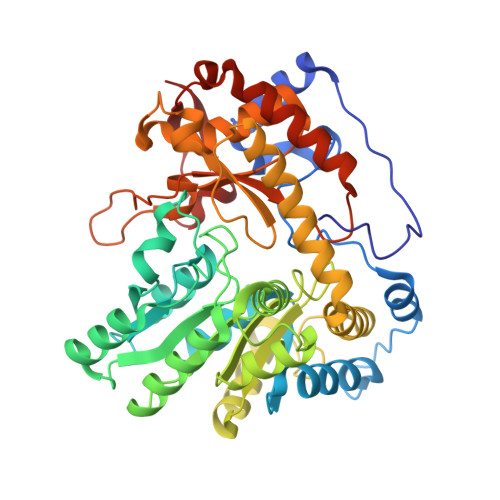
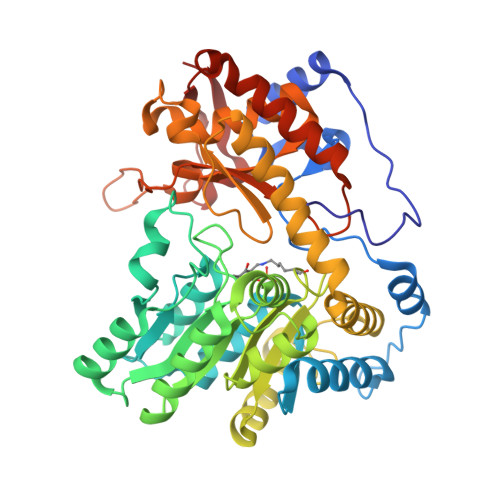
Pressure and Temperature Effects on the Formation of Aminoacrylate Intermediates of Tyrosine Phenol-lyase Demonstrate Reaction Dynamics
Phillips, R.S., Craig, S., Kovalevsky, A., Gerlits, O., Weiss, K., Iorgu, A.I., Heyes, D.J., Hay, S.(2020) ACS Catal 10: 1692-1703