Synthesis of montbretin A analogues yields potent competitive inhibitors of human pancreatic alpha-amylase.
Tysoe, C.R., Caner, S., Calvert, M.B., Win-Mason, A., Brayer, G.D., Withers, S.G.(2019) Chem Sci 10: 11073-11077
- PubMed: 32206255
- DOI: https://doi.org/10.1039/c9sc02610j
- Primary Citation of Related Structures:
6OBX, 6OCN - PubMed Abstract:
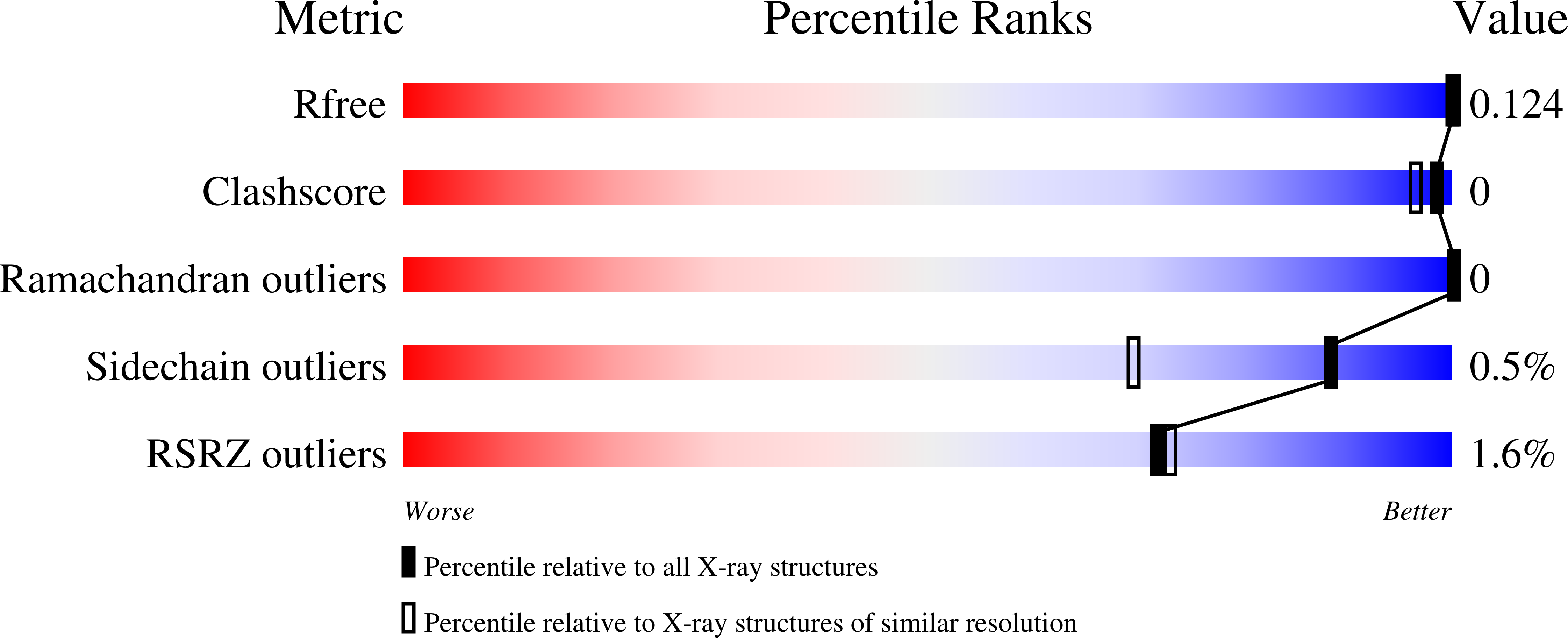
Simplified analogues of the potent human amylase inhibitor montbretin A were synthesised and shown to bind tightly, K I = 60 and 70 nM, with improved specificity over medically relevant glycosidases, making them promising candidates for controlling blood glucose. Crystallographic analysis confirmed similar binding modes and identified new active site interactions.
Organizational Affiliation:
Department of Chemistry , University of British Columbia , 2036 Main Mall , Vancouver BC , Canada V6T 1Z1 . Email: withers@chem.ubc.ca.