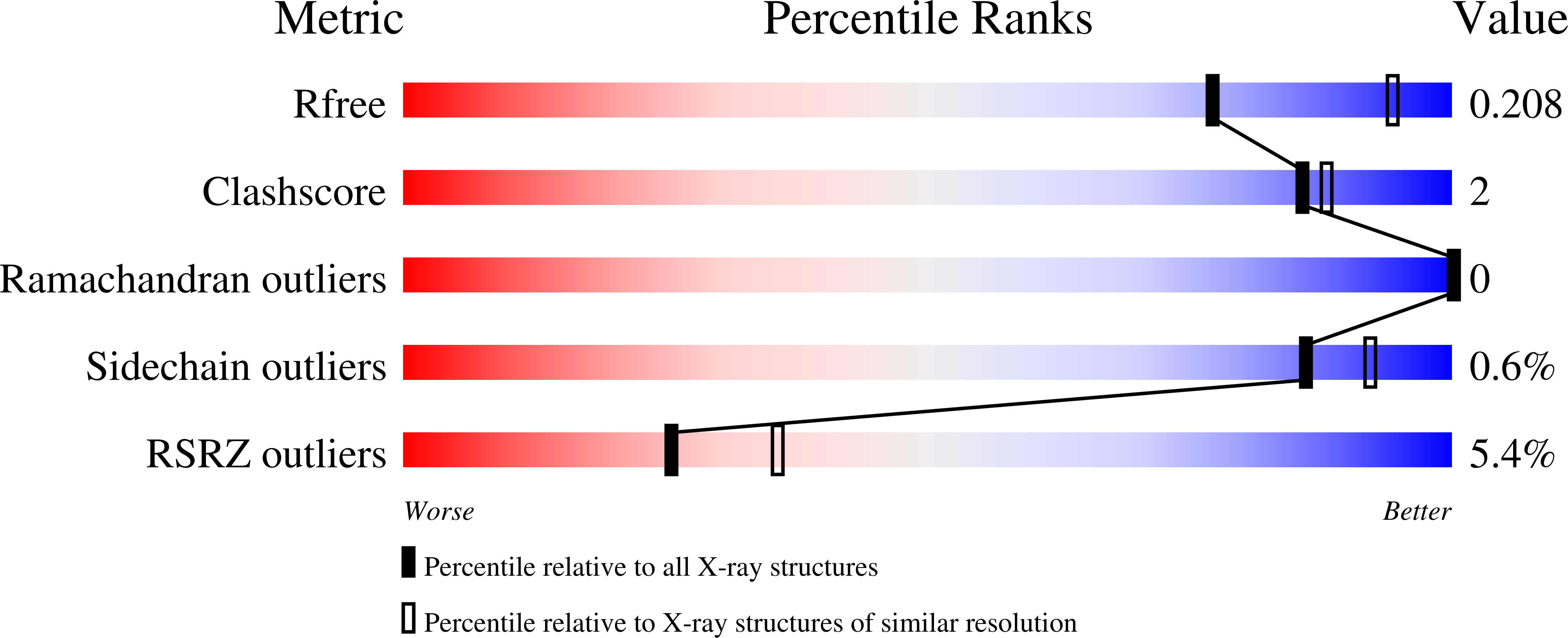
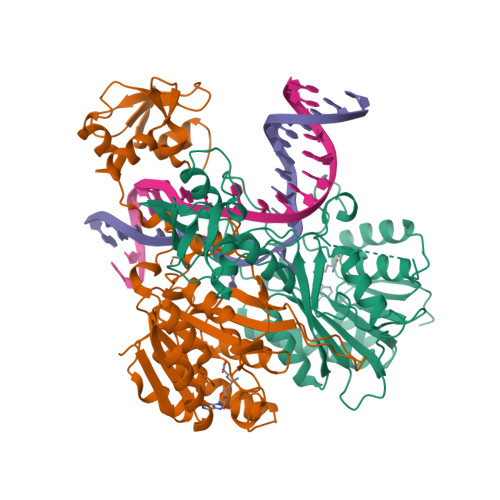
The cell cycle-regulated DNA adenine methyltransferase CcrM opens a bubble at its DNA recognition site.
Horton, J.R., Woodcock, C.B., Opot, S.B., Reich, N.O., Zhang, X., Cheng, X.(2019) Nat Commun 10: 4600-4600
- PubMed: 31601797
- DOI: https://doi.org/10.1038/s41467-019-12498-7
- Primary Citation of Related Structures:
6PBD - PubMed Abstract:
The Caulobacter crescentus cell cycle-regulated DNA methyltransferase (CcrM) methylates the adenine of hemimethylated GANTC after replication. Here we present the structure of CcrM in complex with double-stranded DNA containing the recognition sequence. CcrM contains an N-terminal methyltransferase domain and a C-terminal nonspecific DNA-binding domain. CcrM is a dimer, with each monomer contacting primarily one DNA strand: the methyltransferase domain of one molecule binds the target strand, recognizes the target sequence, and catalyzes methyl transfer, while the C-terminal domain of the second molecule binds the non-target strand. The DNA contacts at the 5-base pair recognition site results in dramatic DNA distortions including bending, unwinding and base flipping. The two DNA strands are pulled apart, creating a bubble comprising four recognized base pairs. The five bases of the target strand are recognized meticulously by stacking contacts, van der Waals interactions and specific Watson-Crick polar hydrogen bonds to ensure high enzymatic specificity.
Organizational Affiliation:
Department of Epigenetics and Molecular Carcinogenesis, University of Texas MD Anderson Cancer Center, Houston, TX, 77030, USA.