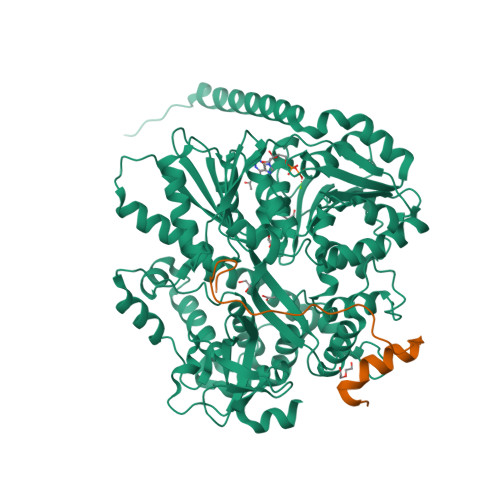
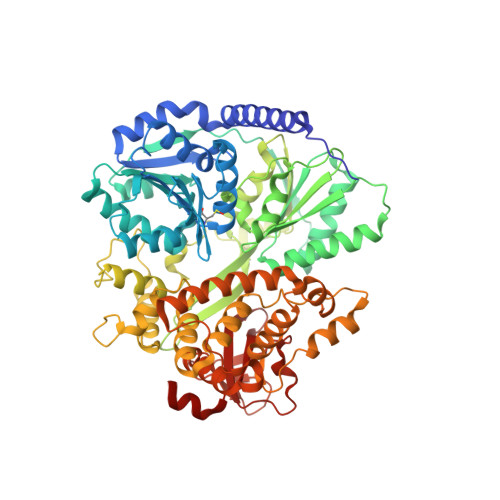
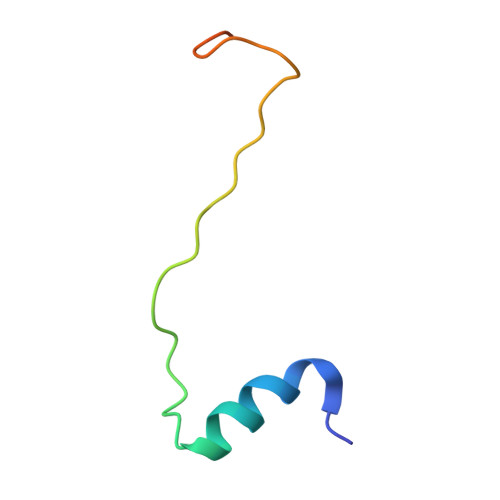
Structural analysis of the intrinsically disordered splicing factor Spp2 and its binding to the DEAH-box ATPase Prp2.
Hamann, F., Schmitt, A., Favretto, F., Hofele, R., Neumann, P., Xiang, S., Urlaub, H., Zweckstetter, M., Ficner, R.(2020) Proc Natl Acad Sci U S A 117: 2948-2956
- PubMed: 31974312
- DOI: https://doi.org/10.1073/pnas.1907960117
- Primary Citation of Related Structures:
6RM8, 6RM9, 6RMA, 6RMB, 6RMC - PubMed Abstract:
The spliceosome consists of five small RNAs and more than 100 proteins. Almost 50% of the human spliceosomal proteins were predicted to be intrinsically disordered or to contain disordered regions, among them the G-patch protein Spp2. The G-patch region of Spp2 binds to the DEAH-box ATPase Prp2, and both proteins together are essential for promoting the transition from the B act to the catalytically active B* spliceosome. Here we show by circular dichroism and nuclear magnetic resonance (NMR) spectroscopy that Spp2 is intrinsically disordered in solution. Crystal structures of a complex consisting of Prp2-ADP and the G-patch domain of Spp2 demonstrate that the G-patch gains a defined fold when bound to Prp2. While the N-terminal region of the G-patch always folds into an α-helix in five different crystal structures, the C-terminal part is able to adopt two alternative conformations. NMR studies further revealed that the N-terminal part of the Spp2 G-patch, which is the most conserved region in different G-patch proteins, transiently samples helical conformations, possibly facilitating a conformational selection binding mechanism. The structural analysis unveils the role of conserved residues of the G-patch in the dynamic interaction mode of Spp2 with Prp2, which is vital to maintain the binding during the Prp2 domain movements needed for RNA translocation.
Organizational Affiliation:
Department of Molecular Structural Biology, Institute of Microbiology and Genetics, Georg-August-University Göttingen, 37077 Göttingen, Germany.