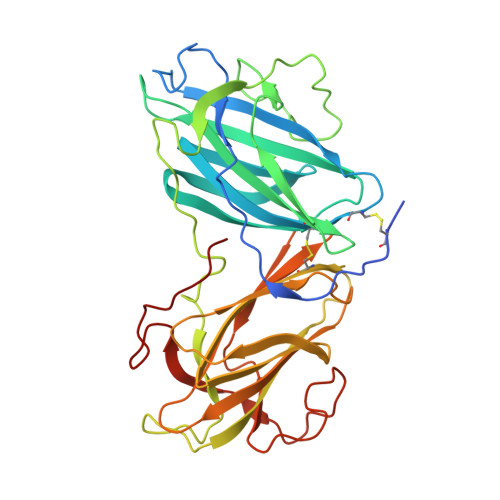
Structure and properties of the oyster mushroom (Pleurotus ostreatus) lectin.
Perduca, M., Destefanis, L., Bovi, M., Galliano, M., Munari, F., Assfalg, M., Ferrari, F., Monaco, H.L., Capaldi, S.(2020) Glycobiology 30: 550-562
- PubMed: 31985778
- DOI: https://doi.org/10.1093/glycob/cwaa006
- Primary Citation of Related Structures:
6T0Q, 6T1D - PubMed Abstract:
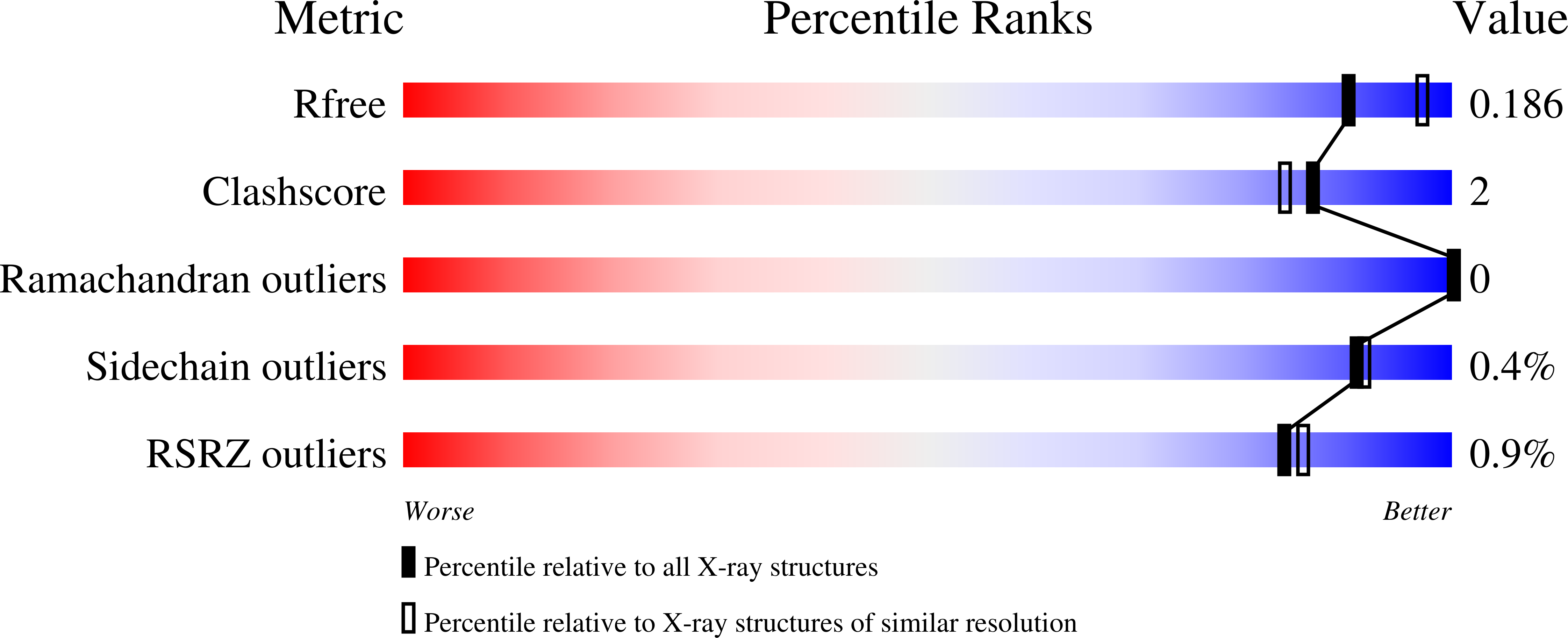
Pleurotus ostreatus Lectin (POL) is a 353 amino acid chain lectin that can be purified from the fruiting bodies of the very well-known and widely diffused edible oyster mushrooms (P. ostreatus). The lectin has been partially characterized by different groups and, although it was crystallized about 20 years ago, its 3D structure and the details of its interactions with carbohydrates are still unknown. This paper reports the 3D structure and ligand-binding properties of POL. We have determined the X-ray structure of the apo-protein purified from the fruiting bodies of the mushroom and that of the recombinant protein in complex with melibiose to a resolution of about 2 Å. The lectin is a homodimer in which the two polypeptide chains are linked by a disulfide bridge. A POL monomer is composed of two highly homologous β-jellyroll domains each of which containing a calcium-dependent carbohydrate-binding site. A high degree of sequence similarity is observed between the two carbohydrate-binding modules present in each monomer. The structure of the lectin in complex with melibiose reveals that a POL dimer has four calcium-dependent carbohydrate-binding sites. The interaction with sugars in solution has been characterized by isothermal titration calorimetry and saturation transfer difference NMR and it sheds new light on the molecular determinants of POL specificity. The lectin exhibits in vitro antiproliferative effects against human cancer cell lines and presents structural similarity with the prototype member of the CBM67 family, the noncatalytic domain of Streptomyces avermitilis α-rhamnosidase.
Organizational Affiliation:
Biocrystallography and Nanostructure Laboratory, Department of Biotechnology, University of Verona, Strada Le Grazie 15, 37134 Verona, Italy.