Enhancing glycan stability via site-selective fluorination: modulating substrate orientation by molecular design.
Axer, A., Jumde, R.P., Adam, S., Faust, A., Schafers, M., Fobker, M., Koehnke, J., Hirsch, A.K.H., Gilmour, R.(2020) Chem Sci 12: 1286-1294
- PubMed: 34163891
- DOI: https://doi.org/10.1039/d0sc04297h
- Primary Citation of Related Structures:
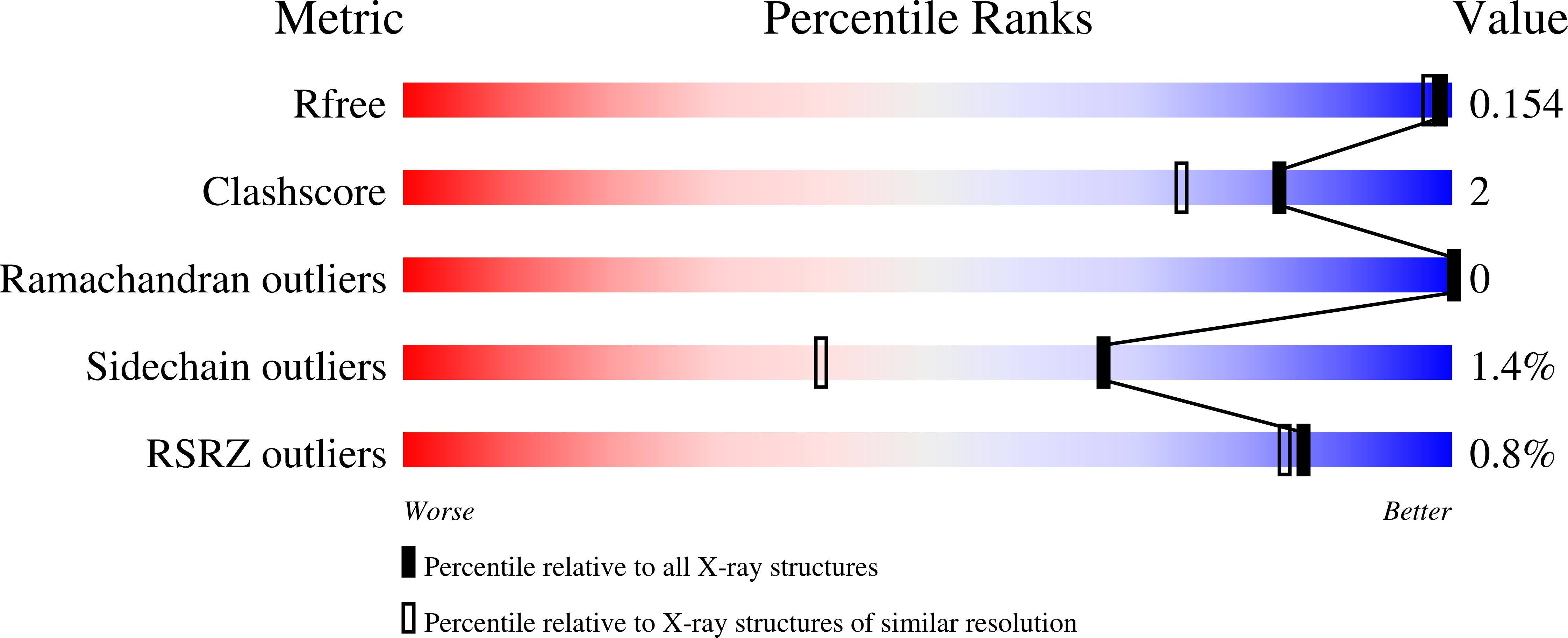
6Z8L - PubMed Abstract:
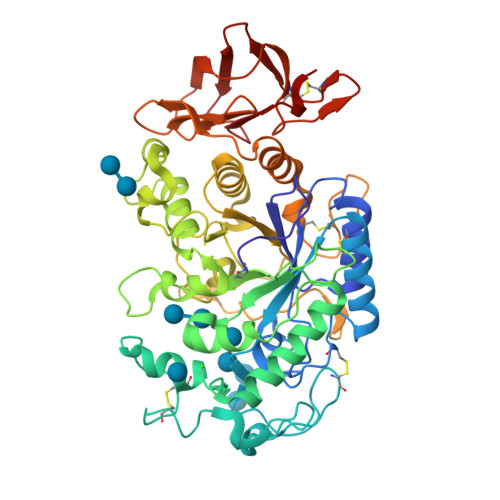
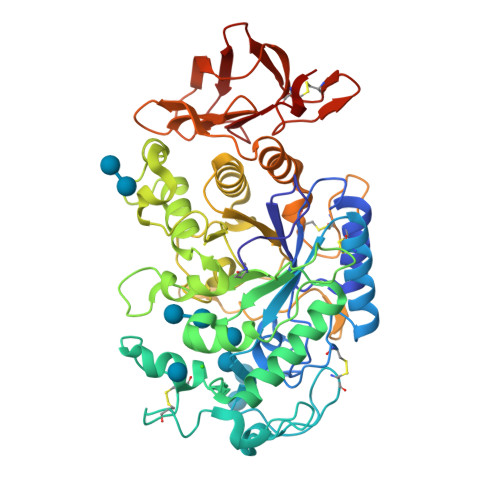
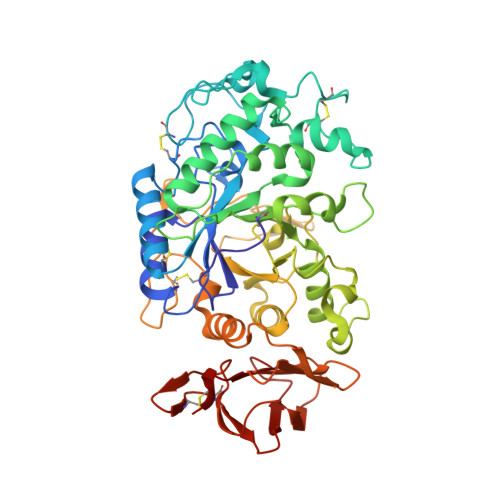
Single site OH → F substitution at the termini of maltotetraose leads to significantly improved hydrolytic stability towards α-amylase and α-glucosidase relative to the natural compound. To explore the effect of molecular editing, selectively modified oligosaccharides were prepared via a convergent α-selective strategy. Incubation experiments in purified α-amylase and α-glucosidase, and in human and murine blood serum, provide insight into the influence of fluorine on the hydrolytic stability of these clinically important scaffolds. Enhancements of ca . 1 order of magnitude result from these subtle single point mutations. Modification at the monosaccharide furthest from the probable enzymatic cleavage termini leads to the greatest improvement in stability. In the case of α-amylase, docking studies revealed that retentive C2-fluorination at the reducing end inverts the orientation in which the substrate is bound. A co-crystal structure of human α-amylase revealed maltose units bound at the active-site. In view of the evolving popularity of C(sp 3 )-F bioisosteres in medicinal chemistry, and the importance of maltodextrins in bacterial imaging, this discovery begins to reconcile the information-rich nature of carbohydrates with their intrinsic hydrolytic vulnerabilities.
Organizational Affiliation:
Organisch Chemisches Institut, WWU Münster Corrensstraße 36 48149 Münster Germany.