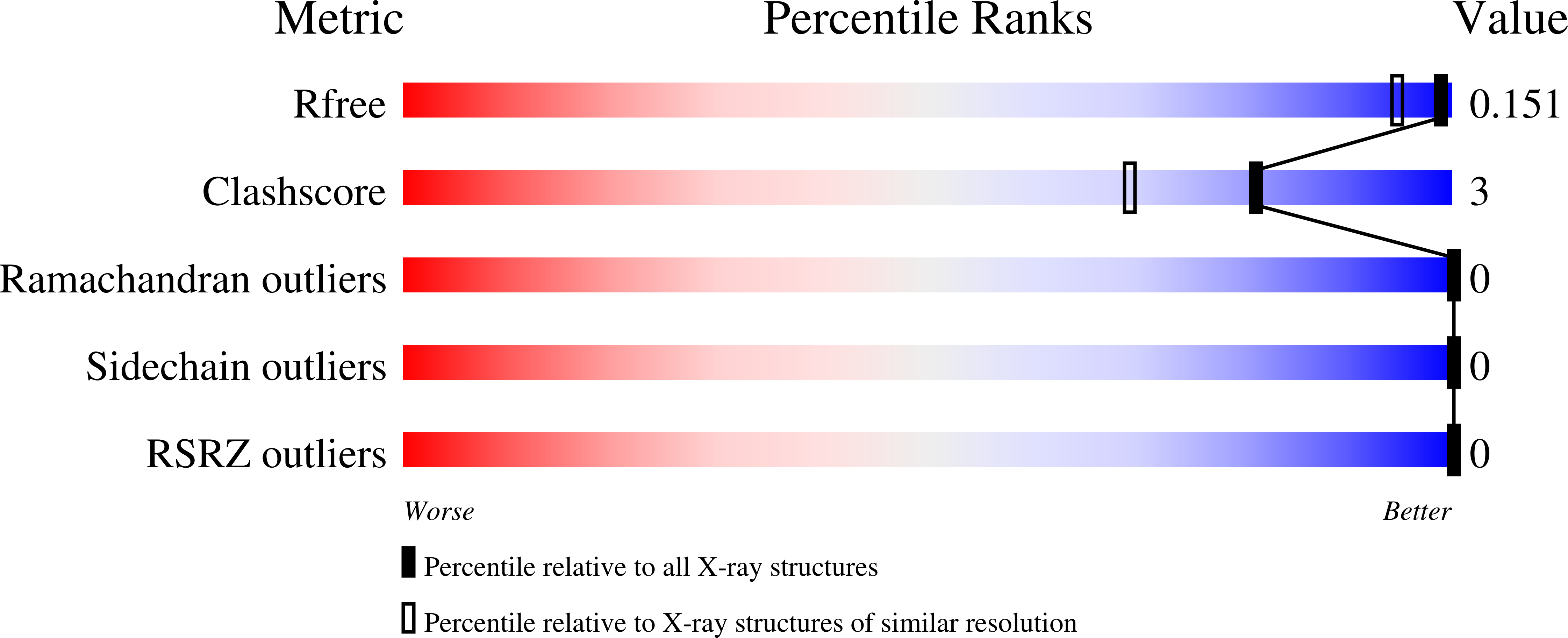
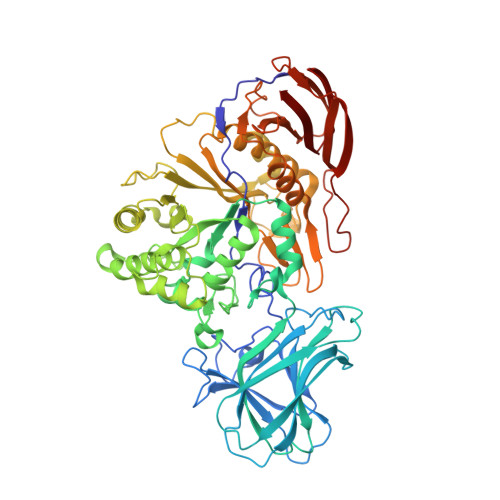
Structure of a GH51 alpha-L-arabinofuranosidase from Meripilus giganteus: conserved substrate recognition from bacteria to fungi.
McGregor, N.G.S., Turkenburg, J.P., Morkeberg Krogh, K.B.R., Nielsen, J.E., Artola, M., Stubbs, K.A., Overkleeft, H.S., Davies, G.J.(2020) Acta Crystallogr D Struct Biol 76: 1124-1133
- PubMed: 33135683
- DOI: https://doi.org/10.1107/S205979832001253X
- Primary Citation of Related Structures:
6ZPS, 6ZPV, 6ZPW, 6ZPX, 6ZPY, 6ZPZ, 6ZQ0, 6ZQ1 - PubMed Abstract:
α-L-Arabinofuranosidases from glycoside hydrolase family 51 use a stereochemically retaining hydrolytic mechanism to liberate nonreducing terminal α-L-arabinofuranose residues from plant polysaccharides such as arabinoxylan and arabinan. To date, more than ten fungal GH51 α-L-arabinofuranosidases have been functionally characterized, yet no structure of a fungal GH51 enzyme has been solved. In contrast, seven bacterial GH51 enzyme structures, with low sequence similarity to the fungal GH51 enzymes, have been determined. Here, the crystallization and structural characterization of MgGH51, an industrially relevant GH51 α-L-arabinofuranosidase cloned from Meripilus giganteus, are reported. Three crystal forms were grown in different crystallization conditions. The unliganded structure was solved using sulfur SAD data collected from a single crystal using the I23 in vacuo diffraction beamline at Diamond Light Source. Crystal soaks with arabinose, 1,4-dideoxy-1,4-imino-L-arabinitol and two cyclophellitol-derived arabinose mimics reveal a conserved catalytic site and conformational itinerary between fungal and bacterial GH51 α-L-arabinofuranosidases.
Organizational Affiliation:
York Structural Biology Laboratory, University of York, Heslington, York YO10 5DD, United Kingdom.