Mechanism of chorismate dehydratase MqnA, the first enzyme of the futalosine pathway, proceeds via substrate-assisted catalysis
Prasad, A., Breithaupt, C., Nguyen, D.A., Lilie, H., Ziegler, J., Stubbs, M.T.(2022) J Biol Chem : 102601
Experimental Data Snapshot
Starting Model: experimental
View more details
(2022) J Biol Chem : 102601
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Chorismate dehydratase | 291 | Streptomyces coelicolor A3(2) | Mutation(s): 1 Gene Names: mqnA, SCO4506 EC: 4.2.1.151 | 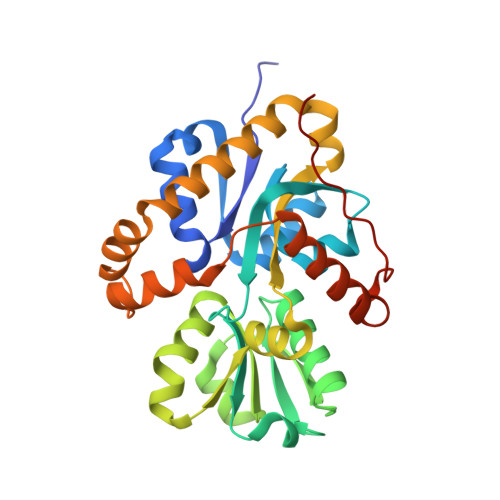 | |
UniProt | |||||
Find proteins for Q9L0T8 (Streptomyces coelicolor (strain ATCC BAA-471 / A3(2) / M145)) Explore Q9L0T8 Go to UniProtKB: Q9L0T8 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | Q9L0T8 | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Ligands 2 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| RDH (Subject of Investigation/LOI) Query on RDH | D [auth A], H [auth B] | 3-[(1-Carboxyvinyl)oxy]benzoic acid C10 H8 O5 HGVAHYJMDVROLE-UHFFFAOYSA-N |  | ||
| GOL Query on GOL | C [auth A] E [auth A] F [auth A] G [auth A] I [auth B] | GLYCEROL C3 H8 O3 PEDCQBHIVMGVHV-UHFFFAOYSA-N |  | ||
| Length ( Å ) | Angle ( ˚ ) |
|---|---|
| a = 44.302 | α = 90 |
| b = 94.778 | β = 95.13 |
| c = 71.472 | γ = 90 |
| Software Name | Purpose |
|---|---|
| PHENIX | refinement |
| XDS | data reduction |
| XSCALE | data scaling |
| PHASER | phasing |
| PDB_EXTRACT | data extraction |
| Funding Organization | Location | Grant Number |
|---|---|---|
| European Regional Development Fund | Germany | -- |