Uncovering a conserved vulnerability site in SARS-CoV-2 by a human antibody.
Li, T., Cai, H., Zhao, Y., Li, Y., Lai, Y., Yao, H., Liu, L.D., Sun, Z., van Vlissingen, M.F., Kuiken, T., GeurtsvanKessel, C.H., Zhang, N., Zhou, B., Lu, L., Gong, Y., Qin, W., Mondal, M., Duan, B., Xu, S., Richard, A.S., Raoul, H., Chen, J., Xu, C., Wu, L., Zhou, H., Huang, Z., Zhang, X., Li, J., Wang, Y., Bi, Y., Rockx, B., Chen, J., Meng, F.L., Lavillette, D., Li, D.(2021) EMBO Mol Med 13: e14544-e14544
- PubMed: 34672091
- DOI: https://doi.org/10.15252/emmm.202114544
- Primary Citation of Related Structures:
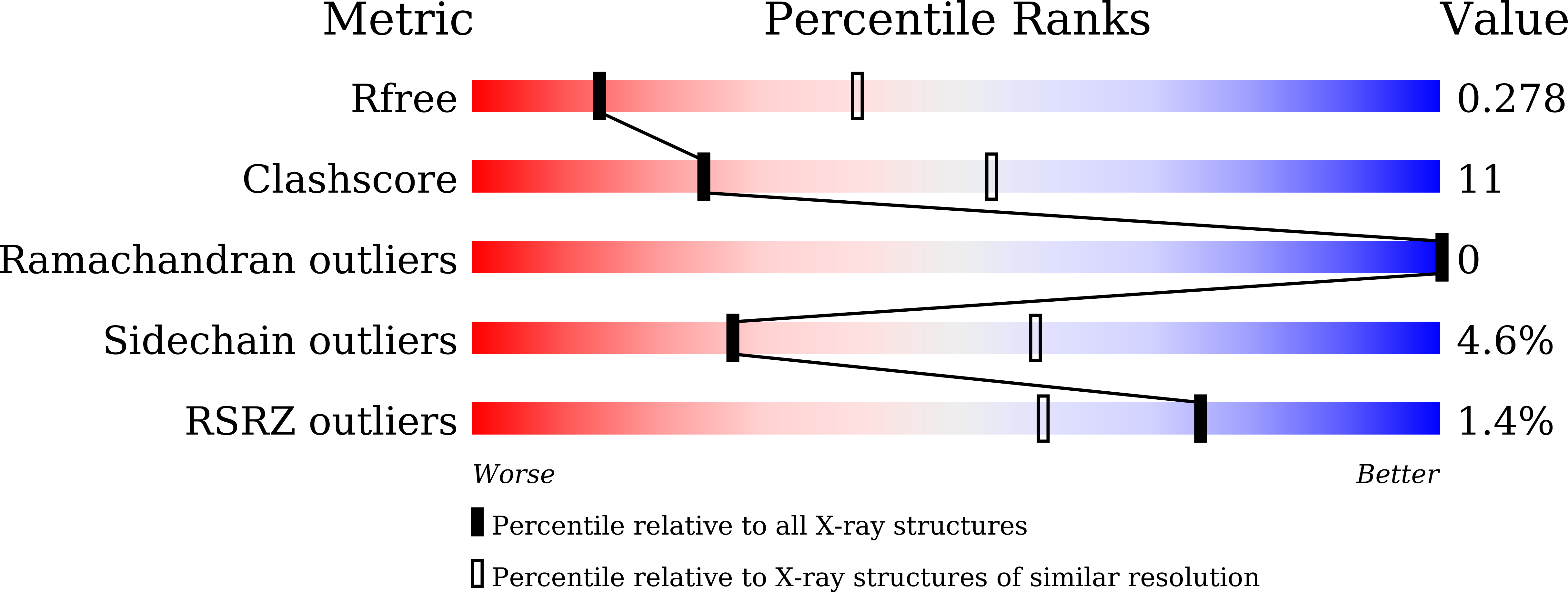
7CYV - PubMed Abstract:
An essential step for SARS-CoV-2 infection is the attachment to the host cell receptor by its Spike receptor-binding domain (RBD). Most of the existing RBD-targeting neutralizing antibodies block the receptor-binding motif (RBM), a mutable region with the potential to generate neutralization escape mutants. Here, we isolated and structurally characterized a non-RBM-targeting monoclonal antibody (FD20) from convalescent patients. FD20 engages the RBD at an epitope distal to the RBM with a K D of 5.6 nM, neutralizes SARS-CoV-2 including the current Variants of Concern such as B.1.1.7, B.1.351, P.1, and B.1.617.2 (Delta), displays modest cross-reactivity against SARS-CoV, and reduces viral replication in hamsters. The epitope coincides with a predicted "ideal" vulnerability site with high functional and structural constraints. Mutation of the residues of the conserved epitope variably affects FD20-binding but confers little or no resistance to neutralization. Finally, in vitro mode-of-action characterization and negative-stain electron microscopy suggest a neutralization mechanism by which FD20 destructs the Spike. Our results reveal a conserved vulnerability site in the SARS-CoV-2 Spike for the development of potential antiviral drugs.
Organizational Affiliation:
State Key Laboratory of Molecular Biology, State Key Laboratory of Cell Biology, CAS Center for Excellence in Molecular Cell Science, Shanghai Institute of Biochemistry and Cell Biology, Chinese Academy of Sciences (CAS), Shanghai, China.