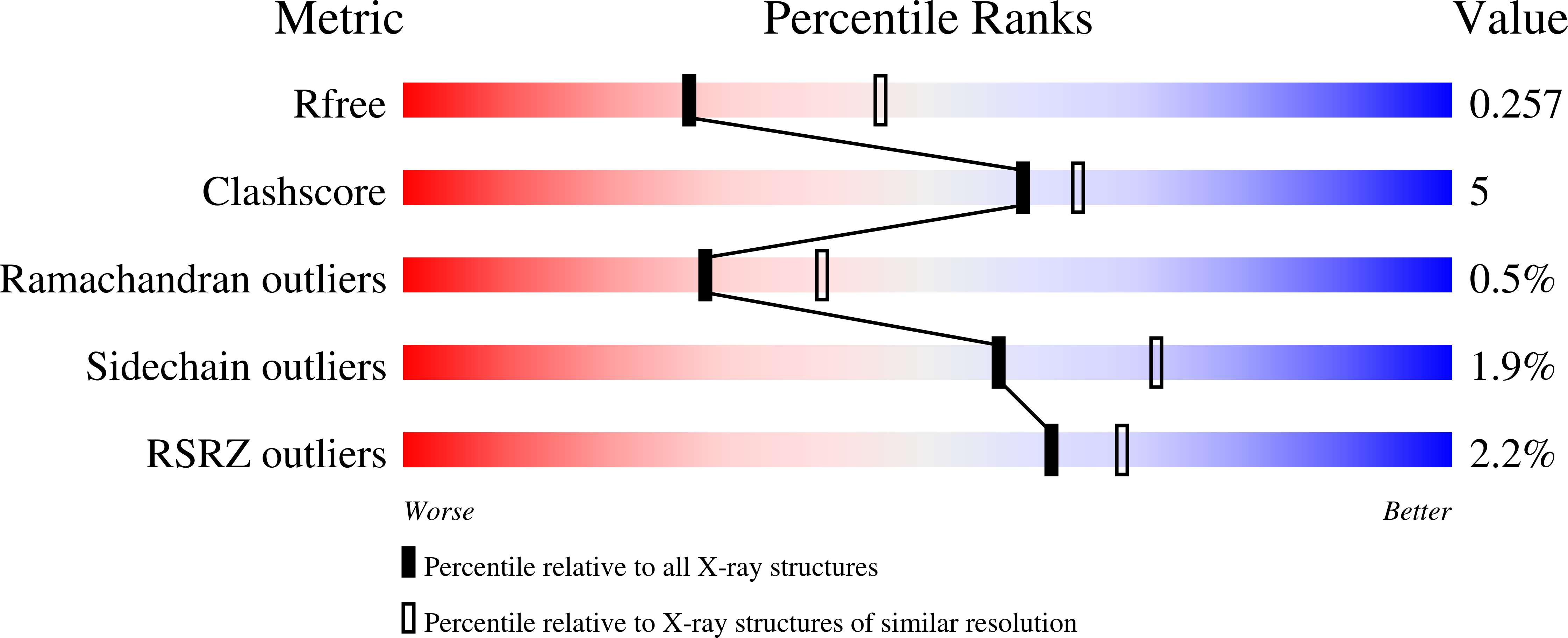
Structural and kinetic investigations of the carboxy terminus of NADPH-cytochrome P450 oxidoreductase.
Hubbard, P.A., Xia, C., Shen, A.L., Kim, J.P.(2021) Arch Biochem Biophys 701: 108792-108792
- PubMed: 33556357
- DOI: https://doi.org/10.1016/j.abb.2021.108792
- Primary Citation of Related Structures:
7L18 - PubMed Abstract:
The influence of the side chains and positioning of the carboxy-terminal residues of NADPH-cytochrome P450 oxidoreductase (CYPOR) on catalytic activity, structure of the carboxy terminus, and interaction with cofactors has been investigated. A tandem deletion of residues Asp675 and Val676, that was expected to shift the position of the functionally important Trp677, resulted in higher cytochrome c reductase activity than that expected from previous studies on the importance of Asp675 and Trp677 in catalysis. Crystallographic determination of the structure of this variant revealed two conformations of the carboxy terminus. In one conformation (Mol A), the last α-helix is partially unwound, resulting in repositioning of all subsequent residues in β-strand 21, from Arg671 to Leu674 (corresponding to Ser673 and Val676 in the wild type structure). This results in the two C-terminal residues, Trp677 and Ser678, being maintained in their wild type positions, with the indole ring of Trp677 stacked against the isoalloxazine ring of FAD as seen in the wild type structure, and Ser673 occupying a similar position to the catalytic residue, Asp675. The other, more disordered conformation is a mixture of the Mol A conformation and one in which the last α-helix is not unwound and the nicotinamide ring is in one of two conformations, out towards the protein surface as observed in the wild type structure (1AMO), or stacked against the flavin ring, similar to that seen in the W677X structure that lacks Trp677 and Ser678 (1JA0). Further kinetic analysis on additional variants showed deletion or substitution of alanine or glycine for Trp677 in conjunction with deletion of Ser678 produced alterations in interactions of CYPOR with NADP + , 2'5'-ADP, and 2'-AMP, as well as the pH dependence of cytochrome c reductase activity. We postulate that deletion of bulky residues at the carboxy terminus permits increased mobility leading to decreased affinity for the 2'5'-ADP and 2'-AMP moieties of NADP + and subsequent domain movement.
Organizational Affiliation:
Department of Biochemistry, Medical College of Wisconsin, Milwaukee, WI, 53226, USA.