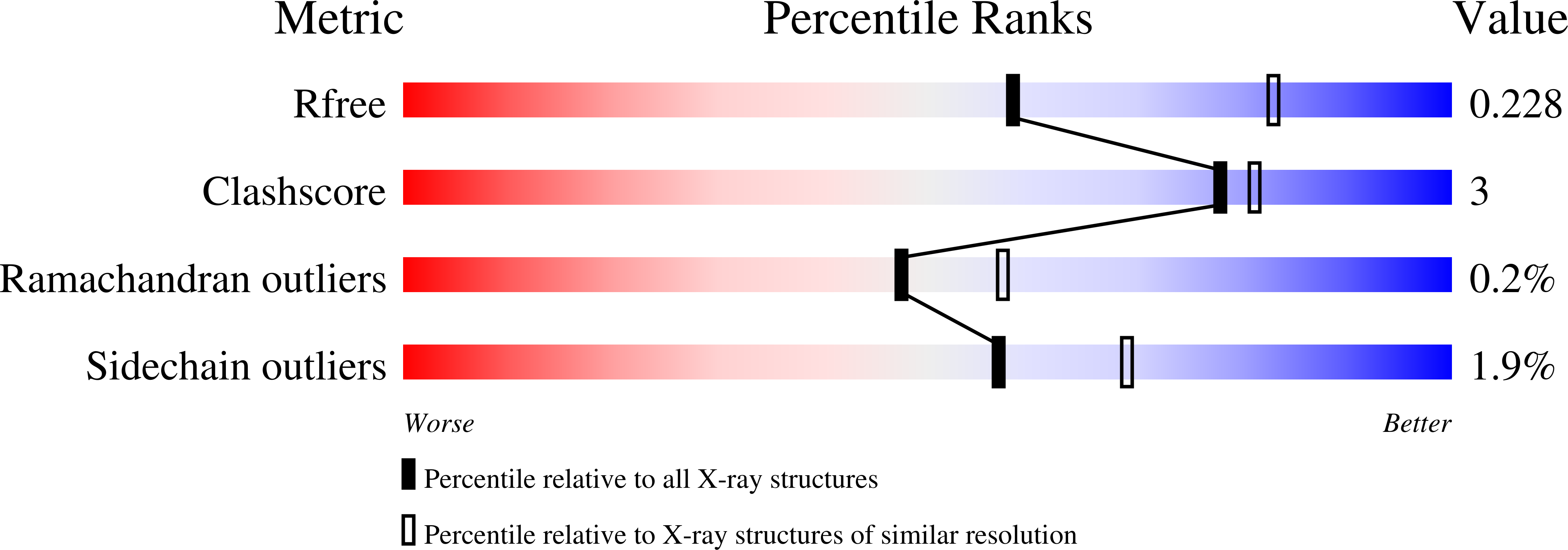
Resistance to the isocitrate dehydrogenase 1 mutant inhibitor ivosidenib can be overcome by alternative dimer-interface binding inhibitors.
Reinbold, R., Hvinden, I.C., Rabe, P., Herold, R.A., Finch, A., Wood, J., Morgan, M., Staudt, M., Clifton, I.J., Armstrong, F.A., McCullagh, J.S.O., Redmond, J., Bardella, C., Abboud, M.I., Schofield, C.J.(2022) Nat Commun 13: 4785-4785
- PubMed: 35970853
- DOI: https://doi.org/10.1038/s41467-022-32436-4
- Primary Citation of Related Structures:
7PJM, 7PJN - PubMed Abstract:
Ivosidenib, an inhibitor of isocitrate dehydrogenase 1 (IDH1) R132C and R132H variants, is approved for the treatment of acute myeloid leukaemia (AML). Resistance to ivosidenib due to a second site mutation of IDH1 R132C, leading to IDH1 R132C/S280F, has emerged. We describe biochemical, crystallographic, and cellular studies on the IDH1 R132C/S280F and R132H/S280F variants that inform on the mechanism of second-site resistance, which involves both modulation of inhibitor binding at the IDH1 dimer-interface and alteration of kinetic properties, which enable more efficient 2-HG production relative to IDH1 R132C and IDH1 R132H. Importantly, the biochemical and cellular results demonstrate that it should be possible to overcome S280F mediated resistance in AML patients by using alternative inhibitors, including some presently in phase 2 clinical trials.
Organizational Affiliation:
Chemistry Research Laboratory, Department of Chemistry and the Ineos Oxford Institute for Antimicrobial Research, University of Oxford, 12 Mansfield, Oxford, OX1 3TA, UK.