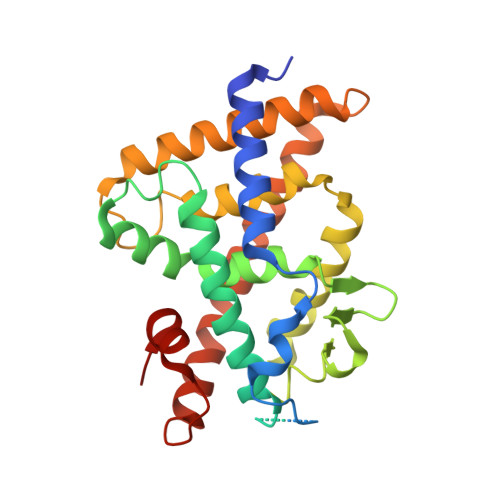
Advances in Vitamin D Receptor Function and Evolution Based on the 3D Structure of the Lamprey Ligand-Binding Domain.
Sigueiro, R., Bianchetti, L., Peluso-Iltis, C., Chalhoub, S., Dejaegere, A., Osz, J., Rochel, N.(2022) J Med Chem 65: 5821-5829
- PubMed: 35302785
- DOI: https://doi.org/10.1021/acs.jmedchem.2c00171
- Primary Citation of Related Structures:
7QPI, 7QPP - PubMed Abstract:
1α,25-dihydroxyvitamin D3 (1,25D 3 ) regulates many physiological processes in vertebrates by binding to the vitamin D receptor (VDR). Phylogenetic analysis indicates that jawless fishes are the most basal vertebrates exhibiting a VDR gene. To elucidate the mechanism driving VDR activation during evolution, we determined the crystal structure of the VDR ligand-binding domain (LBD) complex from the basal vertebrate Petromyzon marinus , sea lamprey (lVDR). Comparison of three-dimensional crystal structures of the lVDR-1,25D 3 complex with higher vertebrate VDR-1,25D 3 structures suggests that 1,25D 3 binds to lVDR similarly to human VDR, but with unique features for lVDR around linker regions between H11 and H12 and between H9 and H10. These structural differences may contribute to the marked species differences in transcriptional responses. Furthermore, residue co-evolution analysis of VDR across vertebrates identifies amino acid positions in H9 and the large insertion domain VDR LBD specific as correlated.
Organizational Affiliation:
Institut de Génétique et de Biologie Moléculaire et Cellulaire (IGBMC), 67400 Illkirch, France.