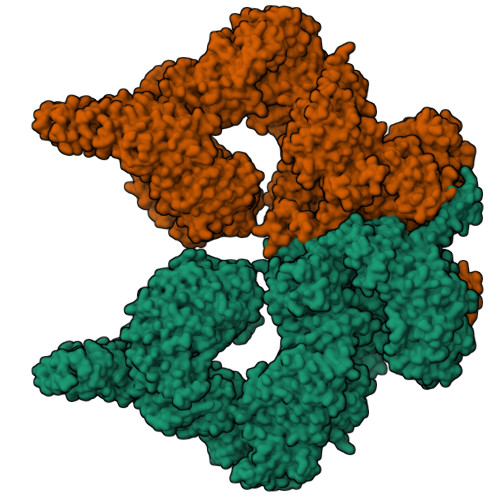
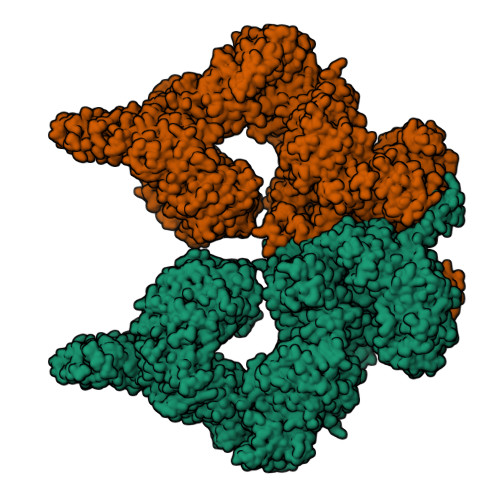
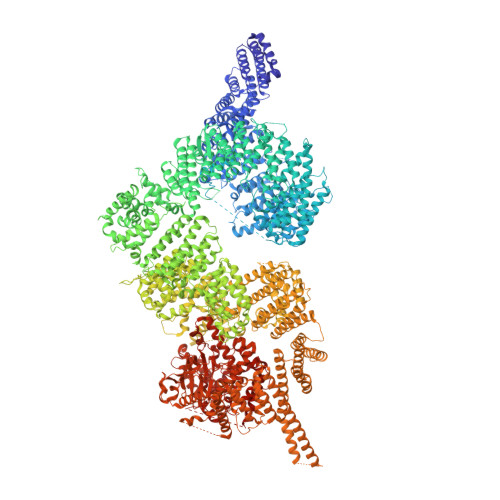
Structure of the human ATM kinase and mechanism of Nbs1 binding.
Warren, C., Pavletich, N.P.(2022) Elife 11
- PubMed: 35076389
- DOI: https://doi.org/10.7554/eLife.74218
- Primary Citation of Related Structures:
7SIC, 7SID - PubMed Abstract:
DNA double-strand breaks (DSBs) can lead to mutations, chromosomal rearrangements, genome instability, and cancer. Central to the sensing of DSBs is the ATM (Ataxia-telangiectasia mutated) kinase, which belongs to the phosphatidylinositol 3-kinase-related protein kinase (PIKK) family. In response to DSBs, ATM is activated by the MRN (Mre11-Rad50-Nbs1) protein complex through a poorly understood process that also requires double-stranded DNA. Previous studies indicate that the FxF/Y motif of Nbs1 directly binds to ATM, and is required to retain active ATM at sites of DNA damage. Here, we report the 2.5 Å resolution cryo-EM structures of human ATM and its complex with the Nbs1 FxF/Y motif. In keeping with previous structures of ATM and its yeast homolog Tel1, the dimeric human ATM kinase adopts a symmetric, butterfly-shaped structure. The conformation of the ATM kinase domain is most similar to the inactive states of other PIKKs, suggesting that activation may involve an analogous realigning of the N and C lobes along with relieving the blockage of the substrate-binding site. We also show that the Nbs1 FxF/Y motif binds to a conserved hydrophobic cleft within the Spiral domain of ATM, suggesting an allosteric mechanism of activation. We evaluate the importance of these structural findings with mutagenesis and biochemical assays.
Organizational Affiliation:
Structural Biology Program, Memorial Sloan Kettering Cancer Center, New York, United States.