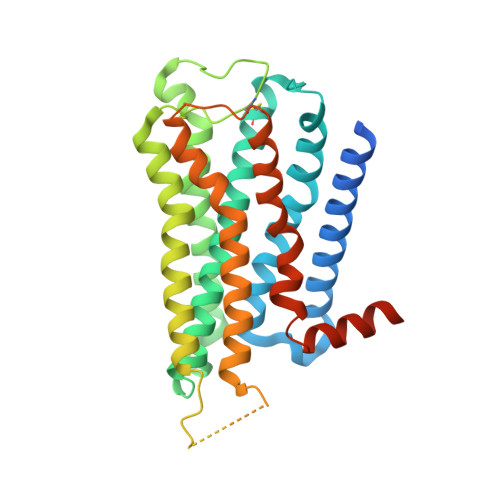
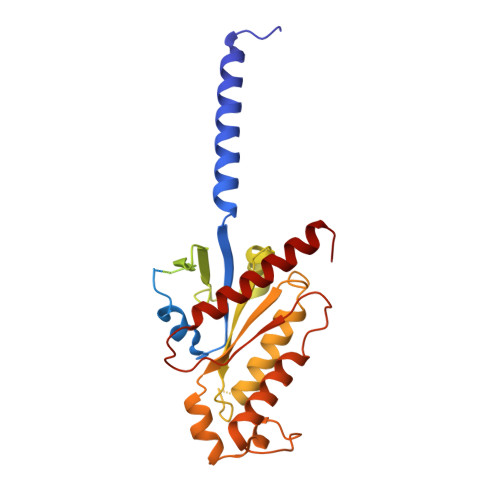
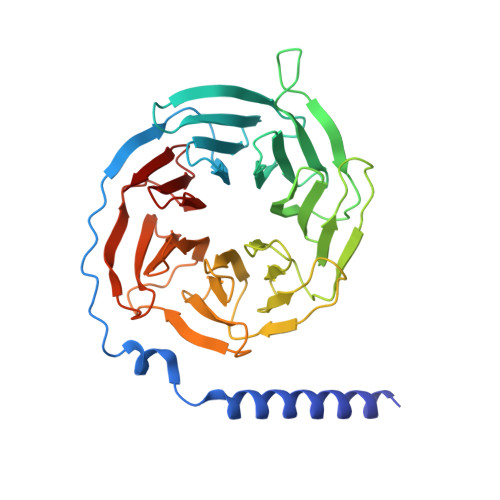
Structural and dynamic insights into supra-physiological activation and allosteric modulation of a muscarinic acetylcholine receptor.
Xu, J., Wang, Q., Hubner, H., Hu, Y., Niu, X., Wang, H., Maeda, S., Inoue, A., Tao, Y., Gmeiner, P., Du, Y., Jin, C., Kobilka, B.K.(2023) Nat Commun 14: 376-376
- PubMed: 36690613
- DOI: https://doi.org/10.1038/s41467-022-35726-z
- Primary Citation of Related Structures:
7T8X, 7T90, 7T94, 7T96 - PubMed Abstract:
The M2 muscarinic receptor (M2R) is a prototypical G-protein-coupled receptor (GPCR) that serves as a model system for understanding GPCR regulation by both orthosteric and allosteric ligands. Here, we investigate the mechanisms governing M2R signaling versatility using cryo-electron microscopy (cryo-EM) and NMR spectroscopy, focusing on the physiological agonist acetylcholine and a supra-physiological agonist iperoxo, as well as a positive allosteric modulator LY2119620. These studies reveal that acetylcholine stabilizes a more heterogeneous M2R-G-protein complex than iperoxo, where two conformers with distinctive G-protein orientations were determined. We find that LY2119620 increases the affinity for both agonists, but differentially modulates agonists efficacy in G-protein and β-arrestin pathways. Structural and spectroscopic analysis suggest that LY211620 stabilizes distinct intracellular conformational ensembles from agonist-bound M2R, which may enhance β-arrestin recruitment while impairing G-protein activation. These results highlight the role of conformational dynamics in the complex signaling behavior of GPCRs, and could facilitate design of better drugs.
Organizational Affiliation:
Department of Molecular and Cellular Physiology, Stanford University School of Medicine, Stanford, CA, 94305, USA.