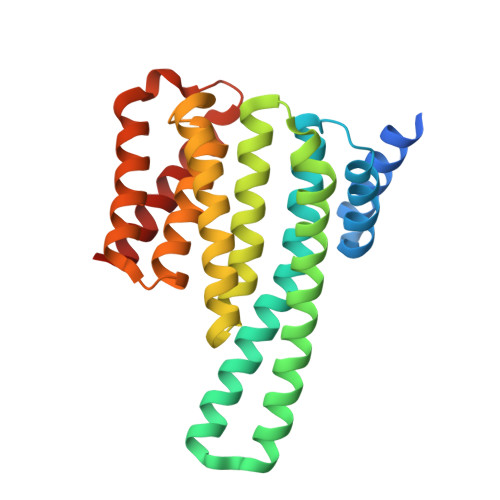
Structure of a 14-3-3 epsilon :FOXO3a pS253 Phosphopeptide Complex Reveals 14-3-3 Isoform-Specific Binding of Forkhead Box Class O Transcription Factor (FOXO) Phosphoproteins.
Mathivanan, S., Chunchagatta Lakshman, P.K., Singh, M., Giridharan, S., Sathish, K., Hurakadli, M.A., Bharatham, K., Kamariah, N.(2022) ACS Omega 7: 24344-24352
- PubMed: 35874228
- DOI: https://doi.org/10.1021/acsomega.2c01700
- Primary Citation of Related Structures:
7V9B - PubMed Abstract:
The transcriptional activity of Forkhead Box O3 (FOXO3a) is inactivated by AKT-mediated phosphorylation on Serine 253 (S253), which enables FOXO3a binding to 14-3-3. Phosphorylated FOXO3a binding to 14-3-3 facilitates the nuclear exclusion of FOXO3a, causing cancer cell proliferation. The FOXO3a/14-3-3 interaction has, therefore, emerged as an important therapeutic target. Here, we report a comprehensive analysis using fluorescence polarization, isothermal titration calorimetry, small-angle X-ray scattering, X-ray crystallography, and molecular dynamics simulations to gain molecular-level insights into the interaction of FOXO3a pS253 phosphopeptide with 14-3-3ε. A high-resolution structure of the fluorophore-labeled FOXO3a pS253 :14-3-3ε complex revealed a distinct mode of interaction compared to other 14-3-3 phosphopeptide complexes. FOXO3a pS253 phosphopeptide showed significant structural difference in the positions of the -3 and -4 Arg residues relative to pSer, compared to that of a similar phosphopeptide, FOXO1 pS256 bound to 14-3-3σ. Moreover, molecular dynamics studies show that the significant structural changes and molecular interactions noticed in the crystal structure of FOXO3a pS253 :14-3-3ε are preserved over the course of the simulation. Thus, this study reveals structural differences between the binding to 14-3-3 isoforms of FOXO1 pS256 versus FOXO3a pS253 , providing a framework for the rational design of isoform-specific FOXO/14-3-3 protein-protein interaction inhibitors for therapy.
Organizational Affiliation:
Centre for Chemical Biology & Therapeutics, inStem & NCBS, Bellary Road, Bangalore 560065, India.