Crystal structures of a 6-dimethylallyltryptophan synthase, IptA: Insights into substrate tolerance and enhancement of prenyltransferase activity.
Suemune, H., Nishimura, D., Mizutani, K., Sato, Y., Hino, T., Takagi, H., Shiozaki-Sato, Y., Takahashi, S., Nagano, S.(2022) Biochem Biophys Res Commun 593: 144-150
- PubMed: 35074664
- DOI: https://doi.org/10.1016/j.bbrc.2022.01.018
- Primary Citation of Related Structures:
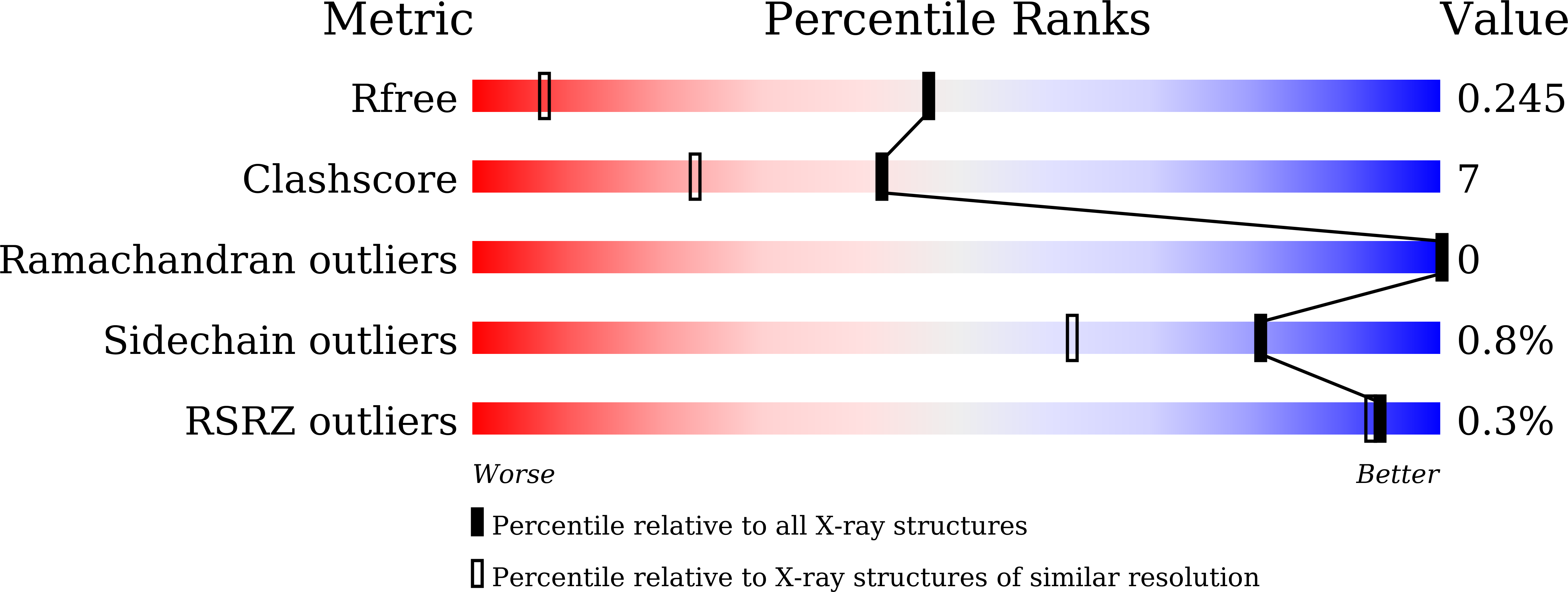
7W8U, 7W8V, 7W8W, 7W8X, 7W8Y - PubMed Abstract:
Dimethylallyltryptophan synthases (DMATSs) catalyze the prenyl transfer reaction from dimethylallyl pyrophosphate (DMAPP) to an indole ring. IptA, a member of the DMATS family, is involved in biosynthesis of 6-dimethylallylindole-3-carbaldehyde in Streptomyces sp. SN-593 and catalyzes the C6-prenylation of l-Trp. The enzyme exhibits prenyl acceptor promiscuity and can accept various Trp derivatives, as observed in several other DMATS family members. Although many crystal structures of DMATS have been determined to date, the structural basis of substrate promiscuity and the acceptance of alternatives to indole-containing natural substrates remain to be clarified. In this study, we determined the crystal structures of the ternary l-Trp derivative (5-methyl-, 6-methyl-, and Nα-methyl-l-Trp) -DMSPP (dimethylallyl S-thiolopyrophosphate; stable analog of DMAPP) -enzyme complex of IptA, in addition to the substrate-free IptA and ternary l-Trp-DMSPP-IptA complex crystal structures. The overall structure of IptA exhibited a typical ABBA-fold, which is commonly found in DMATS family members, while l-Trp and DMSPP are found in a tunnel located inside the ABBA barrel. The crystal structure of the ternary l-Trp-DMSPP-enzyme complex can explain the electrophilic substitution at the C6 atom of l-Trp, which is assisted by Glu84 and His294, as previously suggested for other DMATSs. Although l-Trp snugly fitted into the active site pocket and the unoccupied space around l-Trp is very limited in the l-Trp-DMSPP-IptA complex structure, the enzyme can accommodate 5-methyl- and 6-methyl-l-Trp by slight relocation of the substrate indole ring and adjacent side chain in the active site, resulting in a higher prenylation activity for 5-methyl-l-Trp and C7 prenylation of 6-methyl-l-Trp. Like many other DMATSs, IptA cannot utilize prenyl donors larger than DMAPP. To enlarge the prenyl donor-binding pocket, the W154A mutation was introduced. As expected, this mutant produced prenylated l-Trp from l-Trp and geranyl- and farnesyl pyrophosphate.
Organizational Affiliation:
Department of Chemistry and Biotechnology, Graduate School of Engineering, Tottori University, Tottori, 680-8552, Japan.