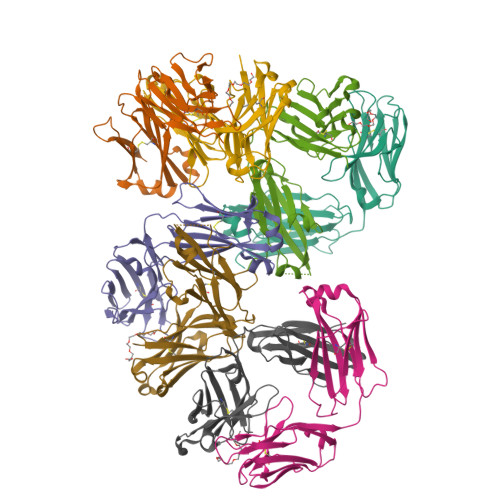
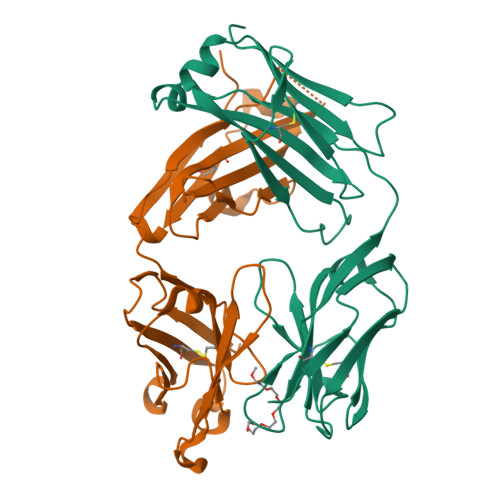
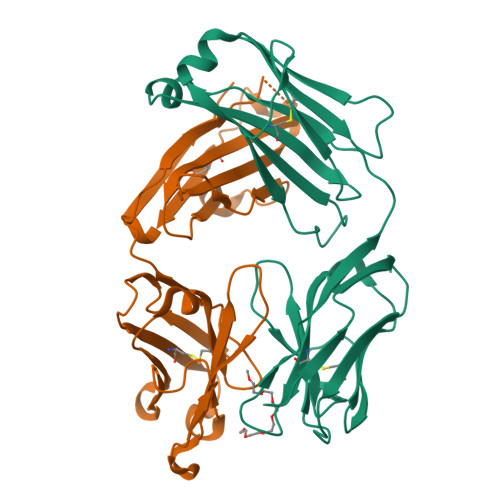
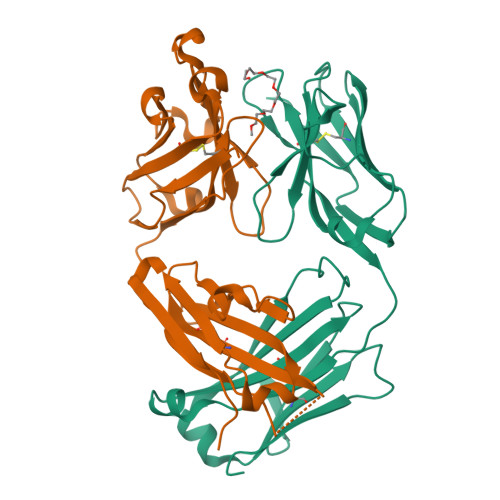
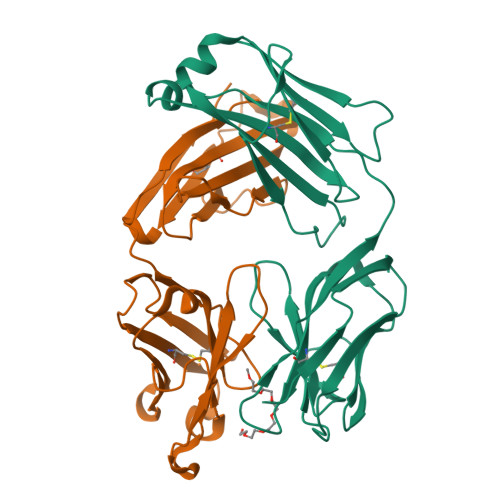
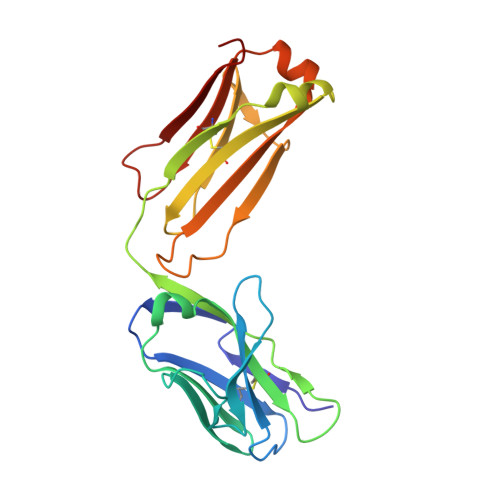
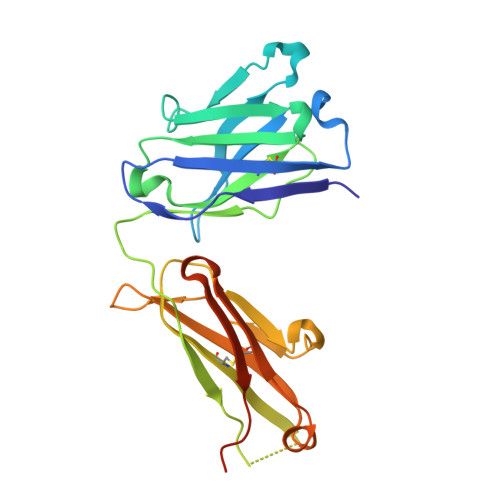
Structural determination of an antibody that specifically recognizes polyethylene glycol with a terminal methoxy group.
Nguyen, M.T., Shih, Y.C., Lin, M.H., Roffler, S.R., Hsiao, C.Y., Cheng, T.L., Lin, W.W., Lin, E.C., Jong, Y.J., Chang, C.Y., Su, Y.C.(2022) Commun Chem 5: 88-88
- PubMed: 35936993
- DOI: https://doi.org/10.1038/s42004-022-00709-0
- Primary Citation of Related Structures:
7Y0G - PubMed Abstract:
Covalent attachment of methoxy poly(ethylene) glycol (mPEG) to therapeutic molecules is widely employed to improve their systemic circulation time and therapeutic efficacy. mPEG, however, can induce anti-PEG antibodies that negatively impact drug therapeutic effects. However, the underlying mechanism for specific binding of antibodies to mPEG remains unclear. Here, we determined the first co-crystal structure of the humanized 15-2b anti-mPEG antibody in complex with mPEG, which possesses a deep pocket in the antigen-binding site to accommodate the mPEG polymer. Structural and mutational analyses revealed that mPEG binds to h15-2b via Van der Waals and hydrogen bond interactions, whereas the methoxy group of mPEG is stabilized in a hydrophobic environment between the V H :V L interface. Replacement of the heavy chain hydrophobic V37 residue with a neutral polar serine or threonine residue offers additional hydrogen bond interactions with methoxyl and hydroxyl groups, resulting in cross-reactivity to mPEG and OH-PEG. Our findings provide insights into understanding mPEG-binding specificity and antigenicity of anti-mPEG antibodies.
Organizational Affiliation:
Department of Biological Science and Technology, Center for Intelligent Drug Systems and Smart Bio-devices (IDS²B), National Yang Ming Chiao Tung University, Hsinchu, Taiwan.