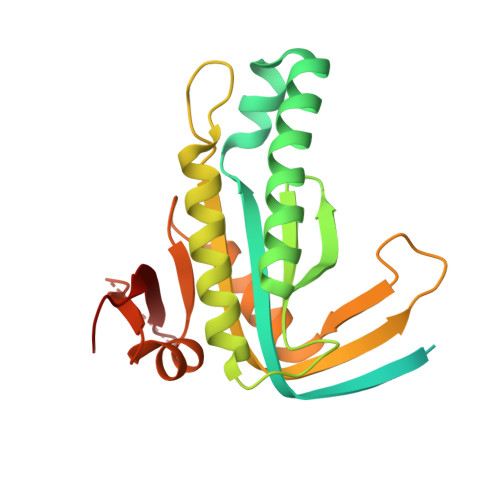
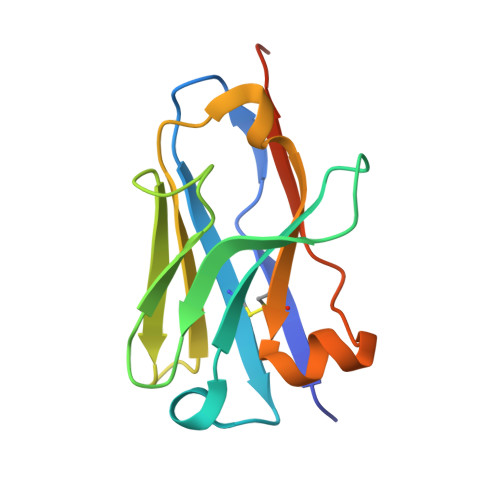
Structure of Mycobacterium tuberculosis Cya, an evolutionary ancestor of the mammalian membrane adenylyl cyclases.
Mehta, V., Khanppnavar, B., Schuster, D., Kantarci, I., Vercellino, I., Kosturanova, A., Iype, T., Stefanic, S., Picotti, P., Korkhov, V.M.(2022) Elife 11
- PubMed: 35980026
- DOI: https://doi.org/10.7554/eLife.77032
- Primary Citation of Related Structures:
7YZ9, 7YZI, 7YZK - PubMed Abstract:
Mycobacterium tuberculosis adenylyl cyclase (AC) Rv1625c/Cya is an evolutionary ancestor of the mammalian membrane ACs and a model system for studies of their structure and function. Although the vital role of ACs in cellular signalling is well established, the function of their transmembrane (TM) regions remains unknown. Here, we describe the cryo-EM structure of Cya bound to a stabilizing nanobody at 3.6 Å resolution. The TM helices 1-5 form a structurally conserved domain that facilitates the assembly of the helical and catalytic domains. The TM region contains discrete pockets accessible from the extracellular and cytosolic side of the membrane. Neutralization of the negatively charged extracellular pocket Ex1 destabilizes the cytosolic helical domain and reduces the catalytic activity of the enzyme. The TM domain acts as a functional component of Cya, guiding the assembly of the catalytic domain and providing the means for direct regulation of catalytic activity in response to extracellular ligands.
Organizational Affiliation:
Laboratory of Biomolecular Research, Division of Biology and Chemistry, Paul Scherrer Institute, Villigen, Switzerland.