Inhibitors against Two PDZ Domains of MDA-9 Suppressed Migration of Breast Cancer Cells.
Tang, H., Wang, L., Li, S., Wei, X., Lv, M., Zhong, F., Liu, Y., Liu, J., Fu, B., Zhu, Q., Wang, D., Liu, J., Ruan, K., Gao, J., Xu, W.(2023) Int J Mol Sci 24
- PubMed: 36834839
- DOI: https://doi.org/10.3390/ijms24043431
- Primary Citation of Related Structures:
8HCK - PubMed Abstract:
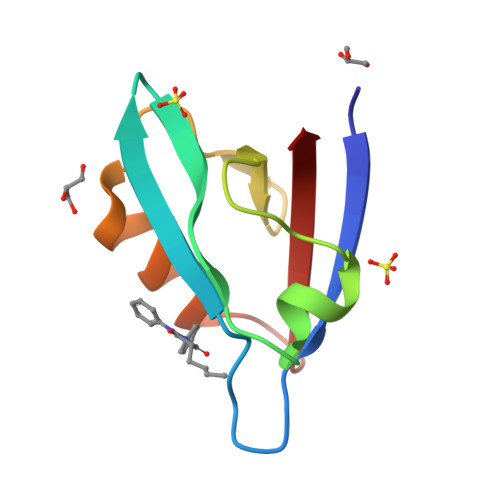
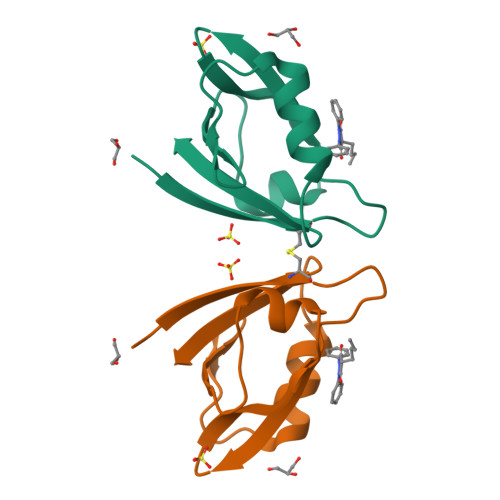
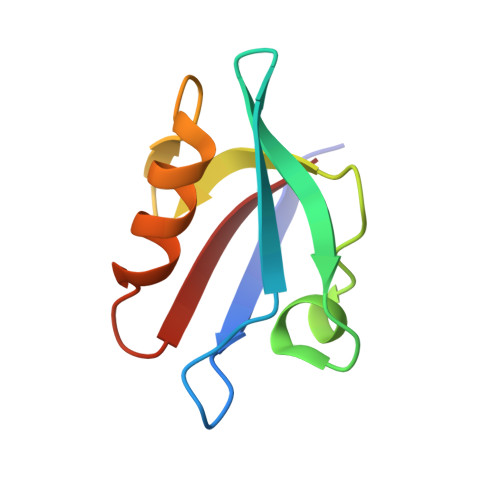
Melanoma differentiation-associated gene 9 (MDA-9) is a small adaptor protein with tandem PDZ domains that promotes tumor progression and metastasis in various human cancers. However, it is difficult to develop drug-like small molecules with high affinity due to the narrow groove of the PDZ domains of MDA-9. Herein, we identified four novel hits targeting the PDZ1 and PDZ2 domains of MDA-9, namely PI1A, PI1B, PI2A, and PI2B, using a protein-observed nuclear magnetic resonance (NMR) fragment screening method. We also solved the crystal structure of the MDA-9 PDZ1 domain in complex with PI1B and characterized the binding poses of PDZ1-PI1A and PDZ2-PI2A, guided by transferred paramagnetic relaxation enhancement. The protein-ligand interaction modes were then cross-validated by the mutagenesis of the MDA-9 PDZ domains. Competitive fluorescence polarization experiments demonstrated that PI1A and PI2A blocked the binding of natural substrates to the PDZ1 and PDZ2 domains, respectively. Furthermore, these inhibitors exhibited low cellular toxicity, but suppressed the migration of MDA-MB-231 breast carcinoma cells, which recapitulated the phenotype of MDA-9 knockdown. Our work has paved the way for the development of potent inhibitors using structure-guided fragment ligation in the future.
Organizational Affiliation:
Institute of Intelligent Machines, Hefei Institutes of Physical Science, Chinese Academy of Sciences, Hefei 230031, China.