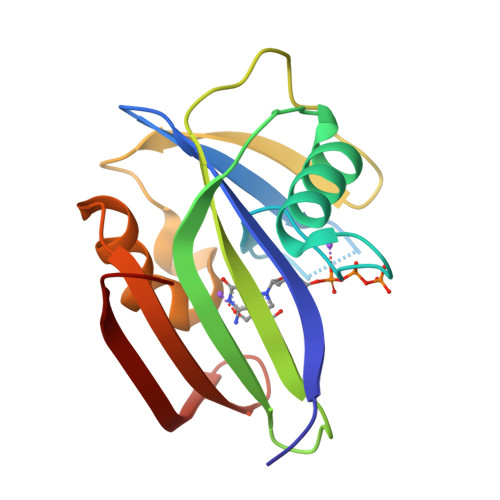
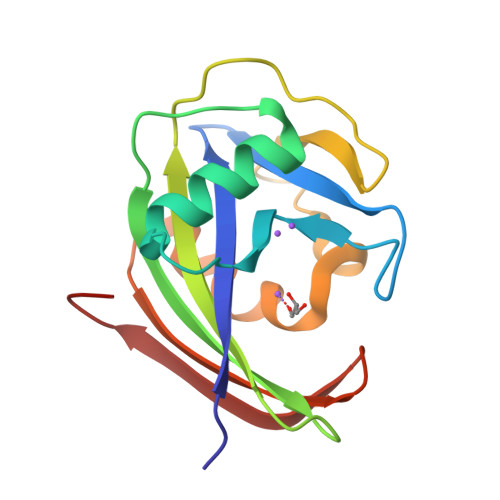
Protonation states of Asp residues in the human Nudix hydrolase MTH1 contribute to its broad substrate recognition.
Nakamura, T., Koga-Ogawa, Y., Fujimiya, K., Chirifu, M., Goto, M., Ikemizu, S., Nakabeppu, Y., Yamagata, Y.(2023) FEBS Lett 597: 1770-1778
- PubMed: 36914375
- DOI: https://doi.org/10.1002/1873-3468.14611
- Primary Citation of Related Structures:
8I18, 8I19, 8I1A, 8I1C, 8I1D, 8I1E, 8I1F, 8I1G, 8I1H, 8I1I, 8I1J, 8I8S, 8I8T - PubMed Abstract:
Human MutT homolog 1 (MTH1), also known as Nudix-type motif 1 (NUDT1), hydrolyzes 8-oxo-dGTP and 2-oxo-dATP with broad substrate recognition and has attracted attention in anticancer therapeutics. Previous studies on MTH1 have proposed that the exchange of the protonation state between Asp119 and Asp120 is essential for the broad substrate recognition of MTH1. To understand the relationship between protonation states and substrate binding, we determined the crystal structures of MTH1 at pH 7.7-9.7. With increasing pH, MTH1 gradually loses its substrate-binding ability, indicating that Asp119 is deprotonated at pH 8.0-9.1 in 8-oxo-dGTP recognition and Asp120 is deprotonated at pH 8.6-9.7 in 2-oxo-dATP recognition. These results confirm that MTH1 recognizes 8-oxo-dGTP and 2-oxo-dATP by exchanging the protonation state between Asp119 and Asp120 with higher pK a .
Organizational Affiliation:
Graduate School of Pharmaceutical Sciences, Kumamoto University, Japan.