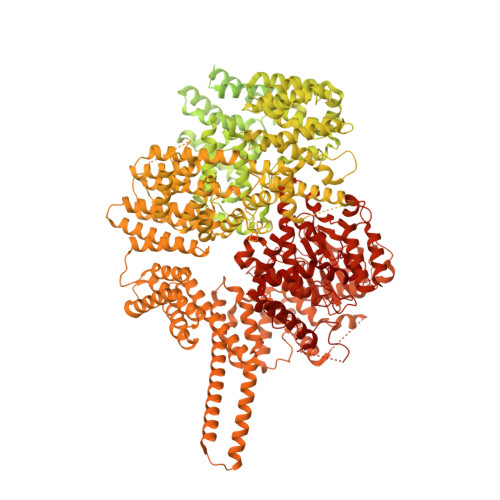
Structural insights into the activation of ataxia-telangiectasia mutated by oxidative stress.
Howes, A.C., Perisic, O., Williams, R.L.(2023) Sci Adv 9: eadi8291-eadi8291
- PubMed: 37756394
- DOI: https://doi.org/10.1126/sciadv.adi8291
- Primary Citation of Related Structures:
8OXM, 8OXO, 8OXP, 8OXQ - PubMed Abstract:
Ataxia-telangiectasia mutated (ATM) is a master kinase regulating DNA damage response that is activated by DNA double-strand breaks. However, ATM is also directly activated by reactive oxygen species, but how oxidative activation is achieved remains unknown. We determined the cryo-EM structure of an H 2 O 2 -activated ATM and showed that under oxidizing conditions, ATM formed an intramolecular disulfide bridge between two protomers that are rotated relative to each other when compared to the basal state. This rotation is accompanied by release of the substrate-blocking PRD region and twisting of the N-lobe relative to the C-lobe, which greatly optimizes catalysis. This active site remodeling enabled us to capture a substrate (p53) bound to the enzyme. This provides the first structural insights into how ATM is activated during oxidative stress.
Organizational Affiliation:
MRC Laboratory of Molecular Biology, Cambridge CB2 0QH, UK.