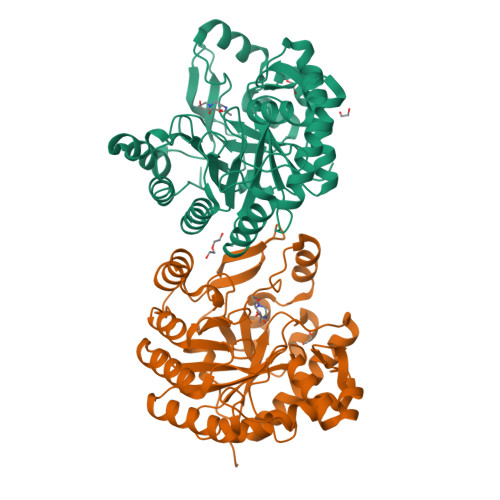
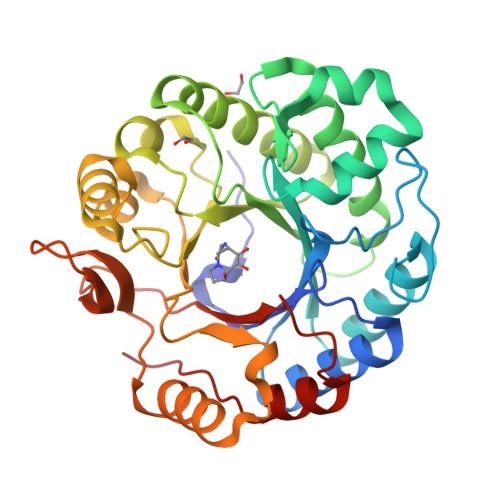
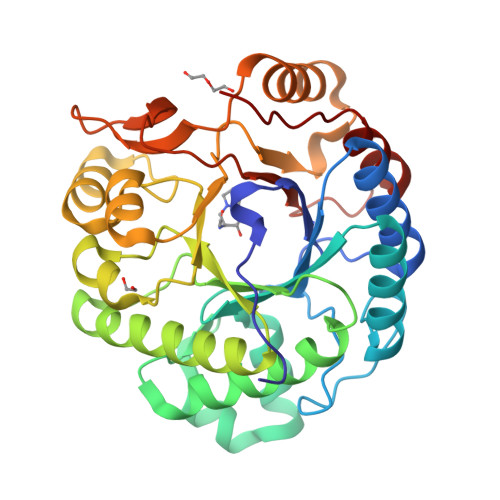
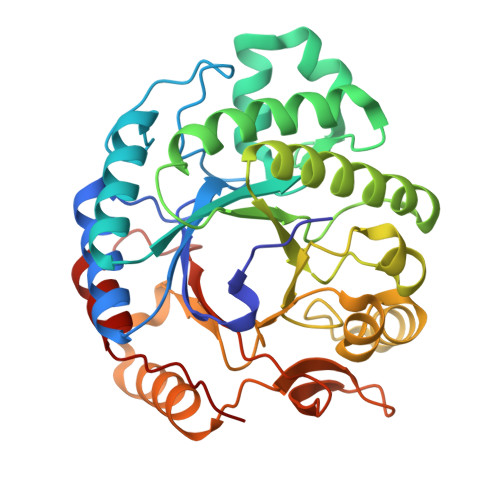
Expansion of the diversity of dispersin scaffolds.
Males, A., Moroz, O.V., Blagova, E., Munch, A., Hansen, G.H., Johansen, A.H., Ostergaard, L.H., Segura, D.R., Eddenden, A., Due, A.V., Gudmand, M., Salomon, J., Sorensen, S.R., Franco Cairo, J.P.L., Nitz, M., Pache, R.A., Vejborg, R.M., Bhosale, S., Vocadlo, D.J., Davies, G.J., Wilson, K.S.(2025) Acta Crystallogr D Struct Biol 81: 130-146
- PubMed: 40019001
- DOI: https://doi.org/10.1107/S205979832500110X
- Primary Citation of Related Structures:
8QAK, 8QB6, 8QCE, 9HTA - PubMed Abstract:
Microorganisms are known to secrete copious amounts of extracellular polymeric substances (EPS) that form complex matrices around the cells to shield them against external stresses, to maintain structural integrity and to influence their environment. Many microorganisms also secrete enzymes that are capable of remodelling or degrading EPS in response to various environmental cues. One key enzyme class is the poly-β-1,6-linked N-acetyl-D-glucosamine (PNAG)-degrading glycoside hydrolases, of which the canonical member is dispersin B (DspB) from CAZy family GH20. We sought to test the hypothesis that PNAG-degrading enzymes would be present across family GH20, resulting in expansion of the sequence and structural space and thus the availability of PNAGases. Phylogenetic analysis revealed that several microorganisms contain potential DspB-like enzymes. Six of these were expressed and characterized, and four crystal structures were determined (two of which were in complex with the established GH20 inhibitor 6-acetamido-6-deoxy-castanospermine and one with a bespoke disaccharide β-1,6-linked thiazoline inhibitor). One enzyme expressed rather poorly, which restricted crystal screening and did not allow activity measurements. Using synthetic PNAG oligomers and MALDI-TOF analysis, two of the five enzymes tested showed preferential endo hydrolytic activity. Their sequences, having only 26% identity to the pioneer enzyme DspB, highlight the considerable array of previously unconsidered dispersins in nature, greatly expanding the range of potential dispersin backbones available for societal application and engineering.
Organizational Affiliation:
York Structural Biology Laboratory, Department of Chemistry, University of York, York YO10 5DD, United Kingdom.