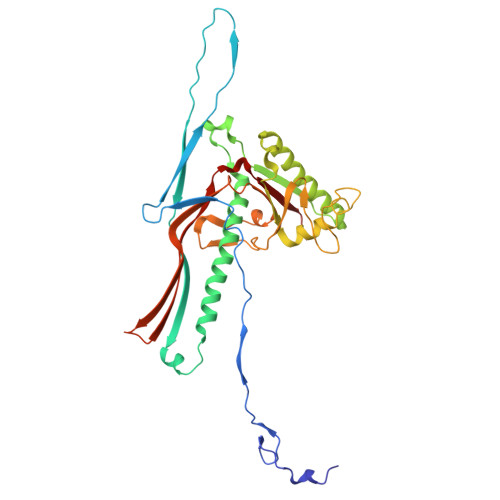
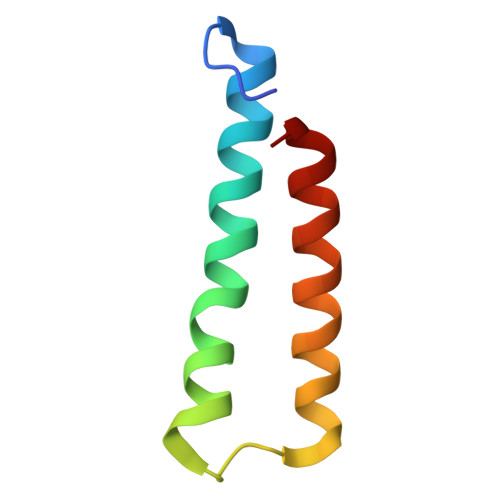
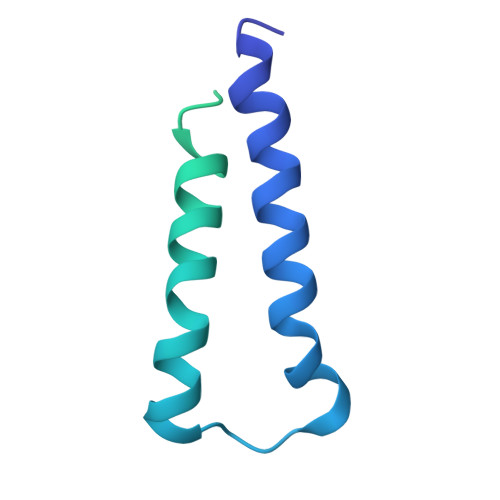
Ejectosome of Pectobacterium bacteriophage Phi M1.
Eruera, A.R., Hodgkinson-Bean, J., Rutter, G.L., Hills, F.R., Kumaran, R., Crowe, A.J.M., Jadav, N., Chang, F., McJarrow-Keller, K., Jorge, F., Hyun, J., Kim, H., Ryu, B., Bostina, M.(2024) PNAS Nexus 3: pgae416-pgae416
- PubMed: 39351541
- DOI: https://doi.org/10.1093/pnasnexus/pgae416
- Primary Citation of Related Structures:
8VB0, 8VB2, 8VB4, 8VBX - PubMed Abstract:
Podophages that infect gram-negative bacteria, such as Pectobacterium pathogen ΦM1, encode tail assemblies too short to extend across the complex gram-negative cell wall. To overcome this, podophages encode a large protein complex (ejectosome) packaged inside the viral capsid and correspondingly ejected during infection to form a transient channel that spans the periplasmic space. Here, we describe the ejectosome of bacteriophage ΦM1 to a resolution of 3.32 Å by single-particle cryo-electron microscopy (cryo-EM). The core consists of tetrameric and octameric ejection proteins which form a ∼1.5-MDa ejectosome that must transition through the ∼30 Å aperture created by the short tail nozzle assembly that acts as the conduit for the passage of DNA during infection. The ejectosome forms several grooves into which coils of genomic DNA are fit before the DNA sharply turns and goes down the tunnel and into the portal. In addition, we reconstructed the icosahedral capsid and hybrid tail apparatus to resolutions between 3.04 and 3.23 Å, and note an uncommon fold adopted by the dimerized decoration proteins which further emphasize the structural diversity of podophages. These reconstructions have allowed the generation of a complete atomic model of the ΦM1, uncovering two distinct decoration proteins and highlighting the exquisite structural diversity of tailed bacteriophages.
Organizational Affiliation:
Department of Microbiology and Immunology, University of Otago, Dunedin 9010, New Zealand.