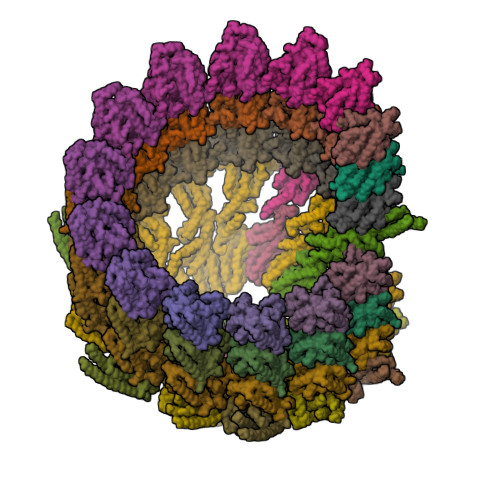
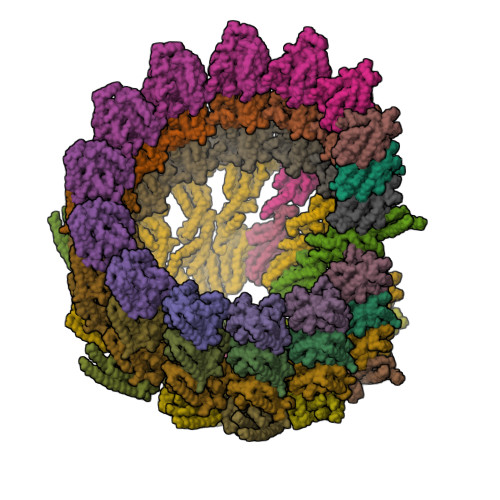
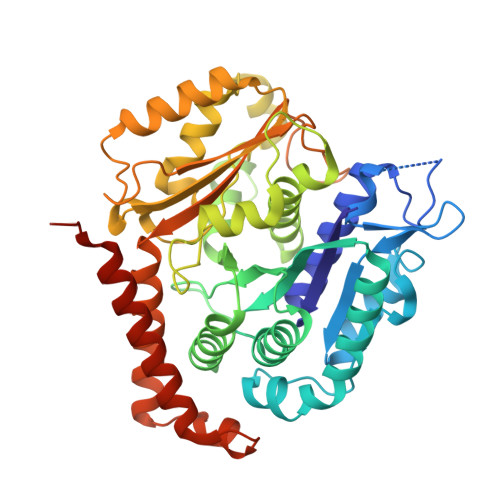
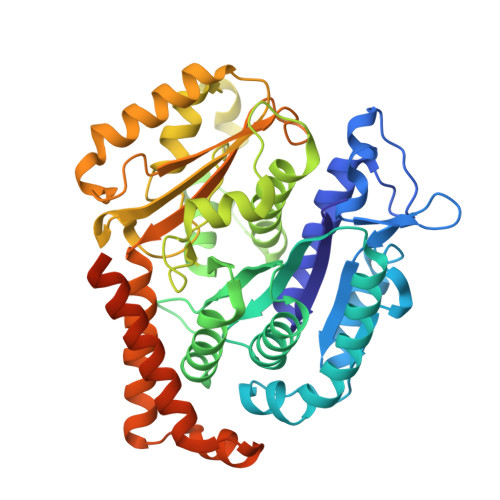
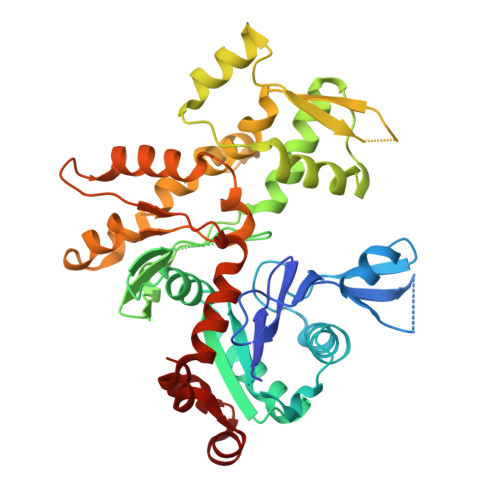
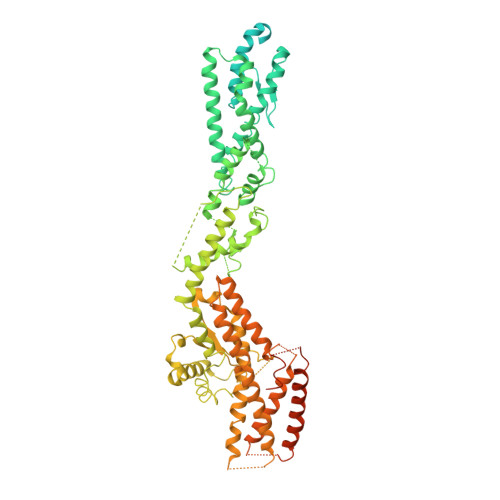
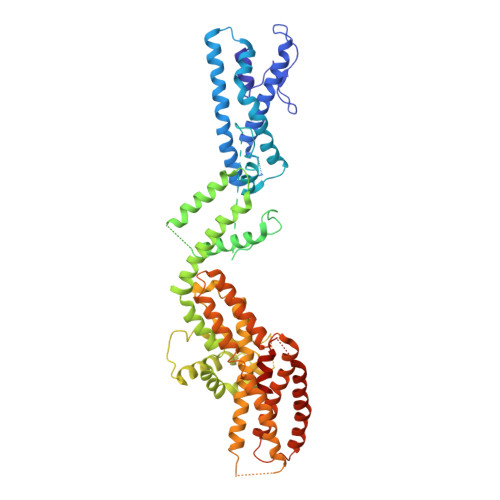
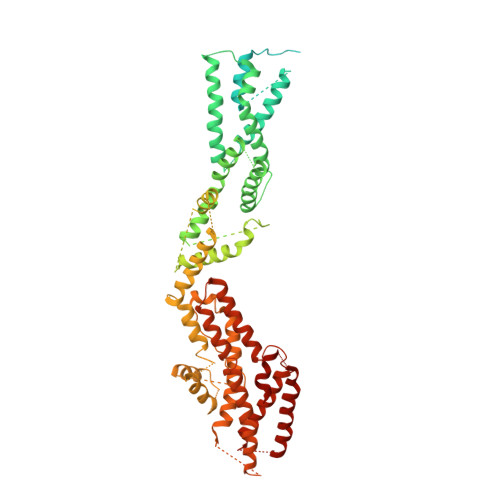
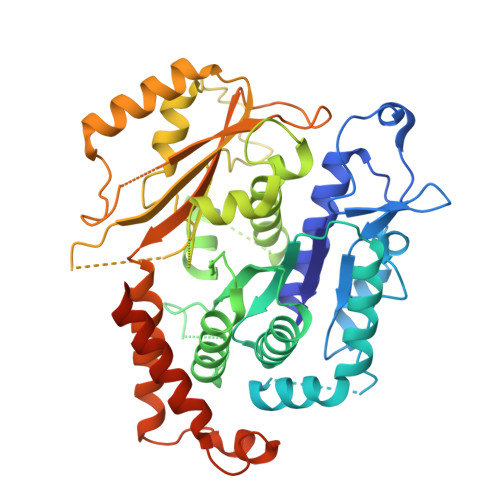
Structure of the gamma-tubulin ring complex-capped microtubule.
Aher, A., Urnavicius, L., Xue, A., Neselu, K., Kapoor, T.M.(2024) Nat Struct Mol Biol 31: 1124-1133
- PubMed: 38609661
- DOI: https://doi.org/10.1038/s41594-024-01264-z
- Primary Citation of Related Structures:
8VA2, 8VRD, 8VRJ, 8VRK, 8VT7 - PubMed Abstract:
Microtubules are composed of α-tubulin and β-tubulin dimers positioned head-to-tail to form protofilaments that associate laterally in varying numbers. It is not known how cellular microtubules assemble with the canonical 13-protofilament architecture, resulting in micrometer-scale α/β-tubulin tracks for intracellular transport that align with, rather than spiral along, the long axis of the filament. We report that the human ~2.3 MDa γ-tubulin ring complex (γ-TuRC), an essential regulator of microtubule formation that contains 14 γ-tubulins, selectively nucleates 13-protofilament microtubules. Cryogenic electron microscopy reconstructions of γ-TuRC-capped microtubule minus ends reveal the extensive intra-domain and inter-domain motions of γ-TuRC subunits that accommodate luminal bridge components and establish lateral and longitudinal interactions between γ-tubulins and α-tubulins. Our structures suggest that γ-TuRC, an inefficient nucleation template owing to its splayed conformation, can transform into a compacted cap at the microtubule minus end and set the lattice architecture of cellular microtubules.
Organizational Affiliation:
Laboratory of Chemistry and Cell Biology, The Rockefeller University, New York, NY, USA.