Mechanistic study of SCOOPs recognition by MIK2-BAK1 complex reveals the role of N-glycans in plant ligand-receptor-coreceptor complex formation
Hu, Y.X., Wu, H.M.(2024) Nature Plants
Experimental Data Snapshot
Starting Model: in silico
View more details
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| MDIS1-interacting receptor like kinase 2 | 683 | Arabidopsis thaliana | Mutation(s): 3 Gene Names: MIK2, At4g08850, T32A17.160 EC: 2.7.11.1 | 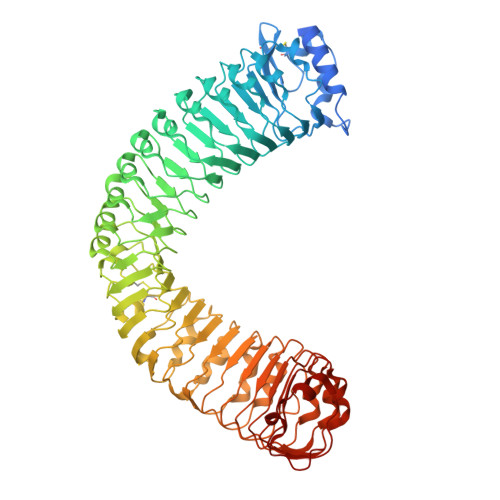 | |
UniProt | |||||
Find proteins for Q8VZG8 (Arabidopsis thaliana) Explore Q8VZG8 Go to UniProtKB: Q8VZG8 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | Q8VZG8 | ||||
Glycosylation | |||||
| Glycosylation Sites: 10 | |||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 2 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| BRASSINOSTEROID INSENSITIVE 1-associated receptor kinase 1 | 230 | Arabidopsis thaliana | Mutation(s): 0 Gene Names: BAK1, ELG, SERK3, At4g33430, F17M5.190 EC: 2.7.10.1 (PDB Primary Data), 2.7.11.1 (PDB Primary Data) | 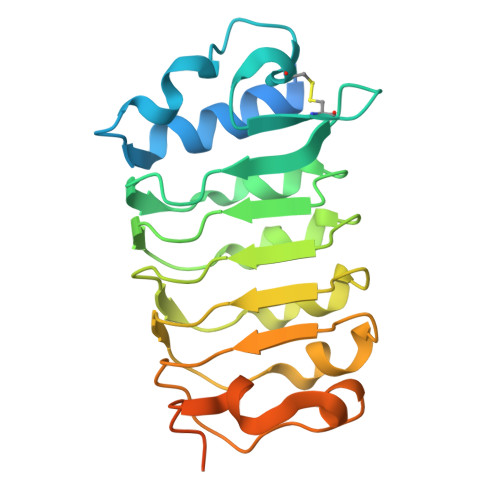 | |
UniProt | |||||
Find proteins for Q94F62 (Arabidopsis thaliana) Explore Q94F62 Go to UniProtKB: Q94F62 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | Q94F62 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Find similar proteins by: Sequence | 3D Structure
Entity ID: 3 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| SERINE-RICH ENDOGENOUS PEPTIDE (SCOOP) | 13 | Arabidopsis thaliana | Mutation(s): 0 | 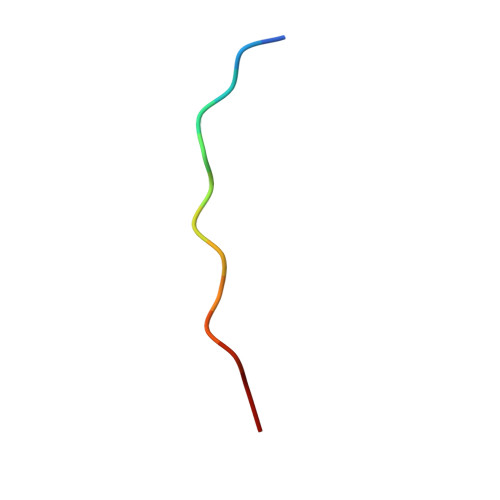 | |
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Ligands 2 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| NAG (Subject of Investigation/LOI) Query on NAG | BA [auth D] CA [auth D] DA [auth D] EA [auth D] FA [auth D] | 2-acetamido-2-deoxy-beta-D-glucopyranose C8 H15 N O6 OVRNDRQMDRJTHS-FMDGEEDCSA-N |  | ||
| BMA (Subject of Investigation/LOI) Query on BMA | AA [auth B] | beta-D-mannopyranose C6 H12 O6 WQZGKKKJIJFFOK-RWOPYEJCSA-N |  | ||
| Length ( Å ) | Angle ( ˚ ) |
|---|---|
| a = 73.004 | α = 90 |
| b = 178.69 | β = 90.79 |
| c = 144.161 | γ = 90 |
| Software Name | Purpose |
|---|---|
| PHENIX | refinement |
| XDS | data reduction |
| Aimless | data scaling |
| PHENIX | phasing |
| Funding Organization | Location | Grant Number |
|---|---|---|
| National Natural Science Foundation of China (NSFC) | China | 32000900 |