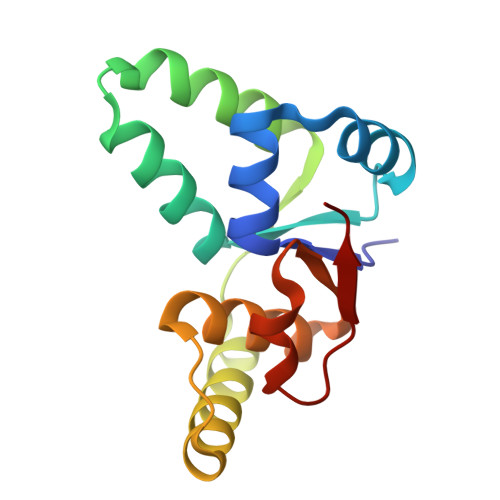
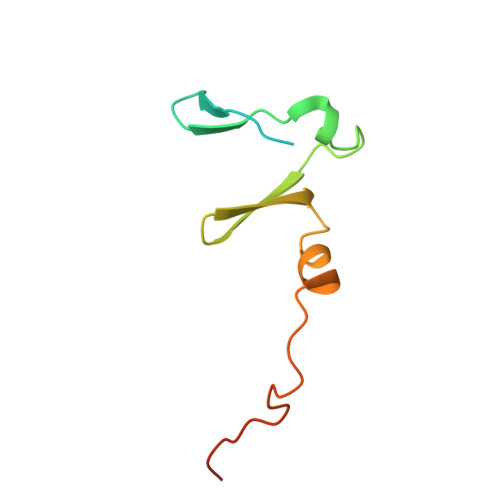
Shared mechanisms of enhanced plasmid maintenance and antibiotic tolerance mediated by the VapBC toxin:antitoxin system
Hollingshead, S., McVicker, G., Nielsen, M.R., Zhang, Y., Pilla, G., Jones, R.A., Thomas, J.C., Johansen, S.E.H., Exley, R.M., Brodersen, D.E., Tang, C.M.To be published.