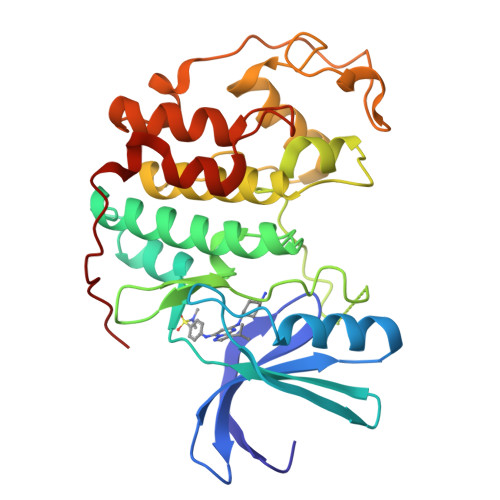
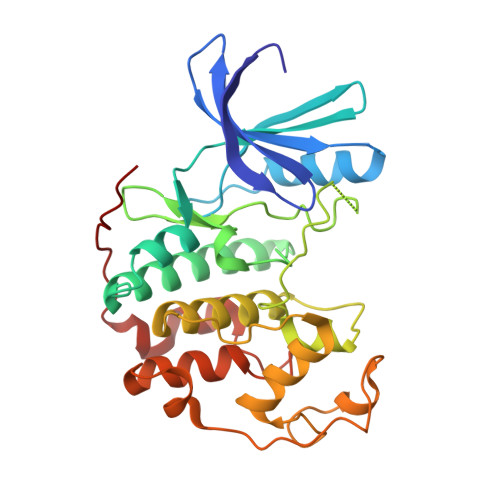
Structure-guided design of pyrazolo[1,5-a]pyrimidines as inhibitors of human cyclin-dependent kinase 2.
Williamson, D.S., Parratt, M.J., Bower, J.F., Moore, J.D., Richardson, C.M., Dokurno, P., Cansfield, A.D., Francis, G.L., Hebdon, R.J., Howes, R., Jackson, P.S., Lockie, A.M., Murray, J.B., Nunns, C.L., Powles, J., Robertson, A., Surgenor, A.E., Torrance, C.J.(2005) Bioorg Med Chem Lett 15: 863-867
- PubMed: 15686876
- DOI: https://doi.org/10.1016/j.bmcl.2004.12.073
- Primary Citation of Related Structures:
1Y8Y, 1Y91 - PubMed Abstract:
The protein structure guided design of a series of pyrazolo[1,5-a]pyrimidines with high potency for human cyclin-dependent kinase 2 (CDK2) is described. Some examples were shown to inhibit the growth of human colon tumour cells, were equipotent for CDK1 and were selective against GSK-3beta and other kinases.
Organizational Affiliation:
Vernalis (R&D) Ltd, Granta Park, Great Abington, Cambridge CB1 6GB, United Kingdom. d.williamson@vernalis.com