Strategic Design of an Effective {beta}-Lactamase Inhibitor: LN-1-255, A 6-ALKYLIDENE-2'-SUBSTITUTED PENICILLIN SULFONE
Pattanaik, P., Bethel, C.R., Hujer, A.M., Hujer, K.M., Distler, A.M., Taracila, M., Anderson, V.E., Fritsche, T.R., Jones, R.N., Pagadala, S.R., van den Akker, F., Buynak, J.D., Bonomo, R.A.(2009) J Biological Chem 284: 945-953
- PubMed: 18955486
- DOI: https://doi.org/10.1074/jbc.M806833200
- Primary Citation of Related Structures:
3D4F - PubMed Abstract:
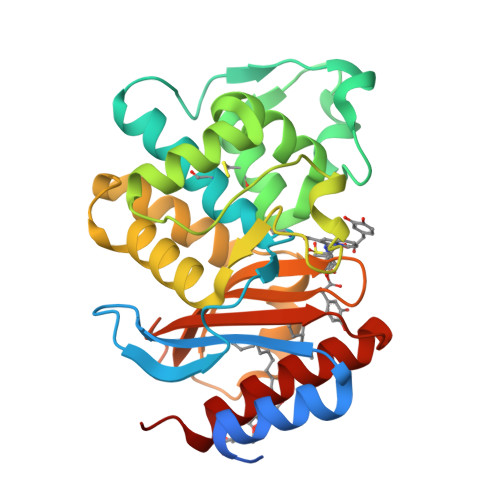
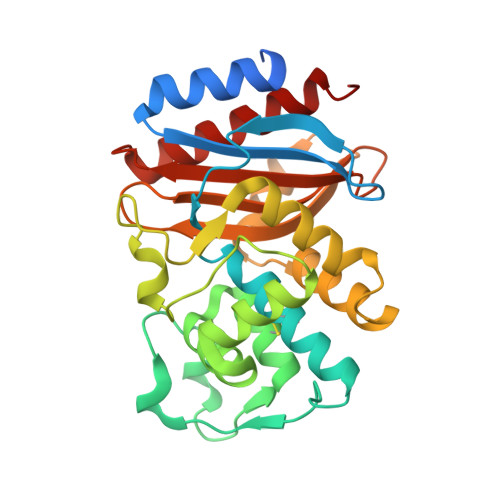
In an effort to devise strategies for overcoming bacterial beta-lactamases, we studied LN-1-255, a 6-alkylidene-2'-substituted penicillin sulfone inhibitor. By possessing a catecholic functionality that resembles a natural bacterial siderophore, LN-1-255 is unique among beta-lactamase inhibitors. LN-1-255 combined with piperacillin was more potent against Escherichia coli DH10B strains bearing bla(SHV) extended-spectrum and inhibitor-resistant beta-lactamases than an equivalent amount of tazobactam and piperacillin. In addition, LN-1-255 significantly enhanced the activity of ceftazidime and cefpirome against extended-spectrum cephalosporin and Sme-1 containing carbapenem-resistant clinical strains. LN-1-255 inhibited SHV-1 and SHV-2 beta-lactamases with nm affinity (K(I) = 110 +/- 10 and 100 +/- 10 nm, respectively). When LN-1-255 inactivated SHV beta-lactamases, a single intermediate was detected by mass spectrometry. The crystal structure of LN-1-255 in complex with SHV-1 was determined at 1.55A resolution. Interestingly, this novel inhibitor forms a bicyclic aromatic intermediate with its carbonyl oxygen pointing out of the oxyanion hole and forming hydrogen bonds with Lys-234 and Ser-130 in the active site. Electron density for the "tail" of LN-1-255 is less ordered and modeled in two conformations. Both conformations have the LN-1-255 carboxyl group interacting with Arg-244, yet the remaining tails of the two conformations diverge. The observed presence of the bicyclic aromatic intermediate with its carbonyl oxygen positioned outside of the oxyanion hole provides a rationale for the stability of this inhibitory intermediate. The 2'-substituted penicillin sulfone, LN-1-255, is proving to be an important lead compound for novel beta-lactamase inhibitor design.
Organizational Affiliation:
Department of Biochemistry, Case Western Reserve University School of Medicine, Cleveland, Ohio 44106, USA.