4-Amino-7,8-dihydro-1,6-naphthyridin-5(6 H)-ones as Inhaled Phosphodiesterase Type 4 (PDE4) Inhibitors: Structural Biology and Structure-Activity Relationships.
Roberts, R.S., Sevilla, S., Ferrer, M., Taltavull, J., Hernandez, B., Segarra, V., Gracia, J., Lehner, M.D., Gavalda, A., Andres, M., Cabedo, J., Vilella, D., Eichhorn, P., Calama, E., Carcasona, C., Miralpeix, M.(2018) J Med Chem 61: 2472-2489
- PubMed: 29502405
- DOI: https://doi.org/10.1021/acs.jmedchem.7b01751
- Primary Citation of Related Structures:
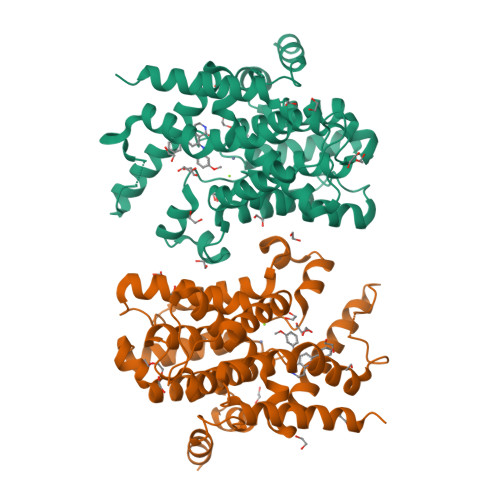
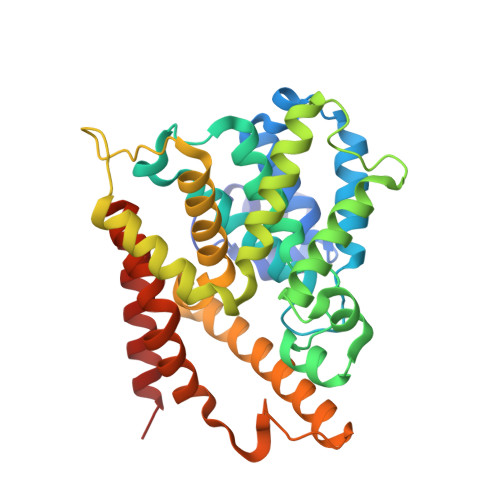
5K32 - PubMed Abstract:
Rational design of a novel template of naphthyridinones rapidly led to PDE4 inhibitors with subnanomolar enzymatic potencies. X-ray crystallography confirmed the binding mode of this novel template. We achieved compounds with double-digit picomolar enzymatic potencies through further structure-based design by targeting both the PDE4 enzyme metal-binding pocket and occupying the solvent-filled pocket. A strategy for lung retention and long duration of action based on low aqueous solubility was followed. In vivo efficacies were measured in a rat lung neutrophilia model by suspension microspray and dry powder administration. Suspension microspray of potent compounds showed in vivo efficacy with a clear dose-response. Despite sustained lung levels, dry powder administration performed much less well and without proper dose-response, highlighting clear differences between the two formulations. This indicates a deficiency in the low aqueous solubility strategy for long duration lung efficacy.
Organizational Affiliation:
Medicinal Chemistry & Screening , ‡Pharmacokinetics & Metabolism , and §Experimental Dermatology , Almirall S.A., Centro de Investigación y Desarrollo , Crta. Laureà Miró 408-410 , Sant Feliu de Llobregat, 08980 Barcelona , Spain.