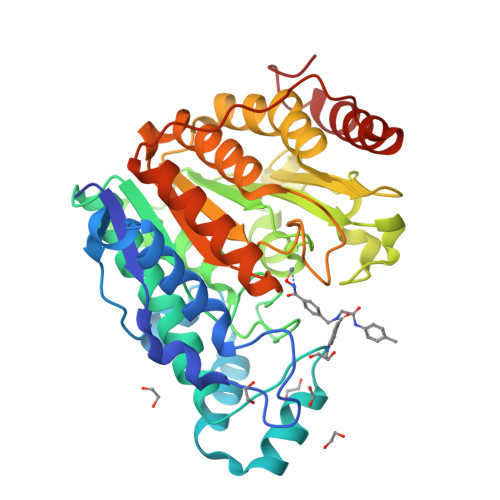
Histone Deacetylase 6-Selective Inhibitors and the Influence of Capping Groups on Hydroxamate-Zinc Denticity.
Porter, N.J., Osko, J.D., Diedrich, D., Kurz, T., Hooker, J.M., Hansen, F.K., Christianson, D.W.(2018) J Med Chem 61: 8054-8060
- PubMed: 30118224
- DOI: https://doi.org/10.1021/acs.jmedchem.8b01013
- Primary Citation of Related Structures:
6DVL, 6DVM, 6DVN, 6DVO - PubMed Abstract:
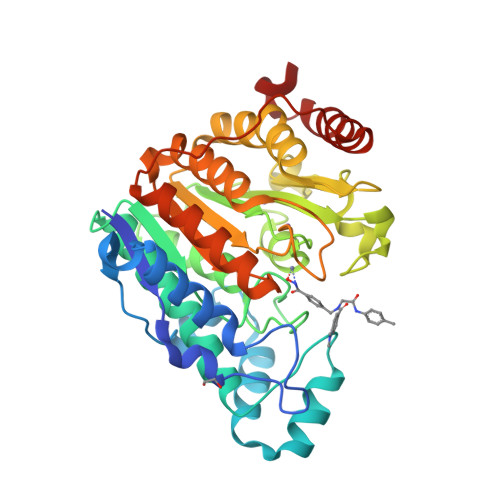
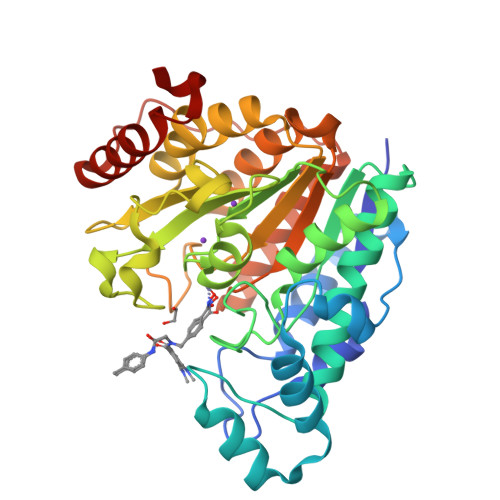
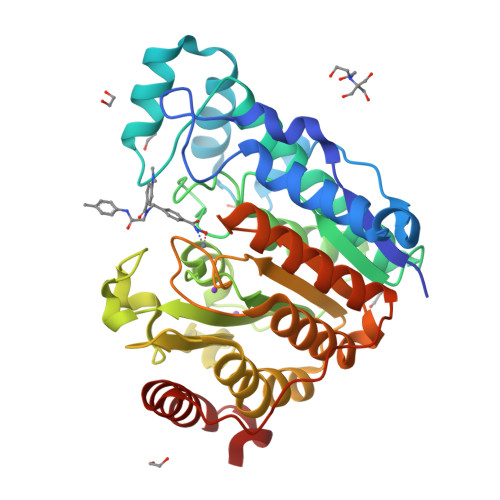
Four crystal structures are presented of histone deacetylase 6 (HDAC6) complexes with para-substituted phenylhydromaxamate inhibitors, including bulky peptoids. These structures provide insight regarding the design of capping groups that confer selectivity for binding to HDAC6, specifically with regard to interactions in a pocket formed by the L1 loop. Capping group interactions may also influence hydroxamate-Zn 2+ coordination with monodentate or bidentate geometry.
Organizational Affiliation:
Roy and Diana Vagelos Laboratories, Department of Chemistry , University of Pennsylvania , 231 South 34th Street , Philadelphia , Pennsylvania 19104-6323 , United States.