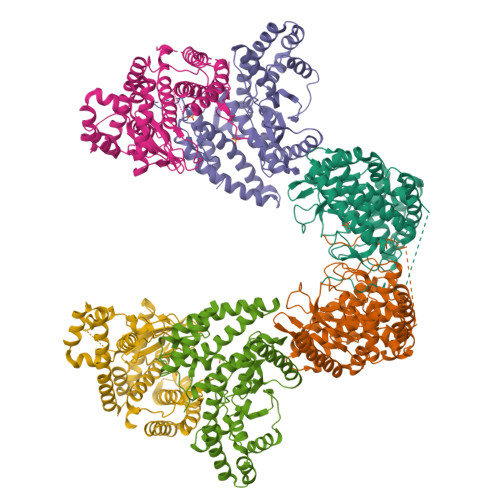
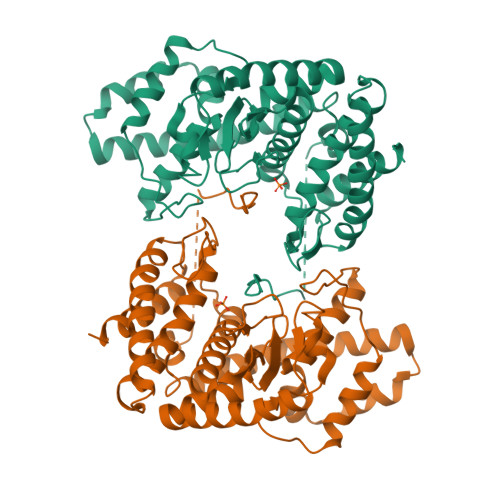
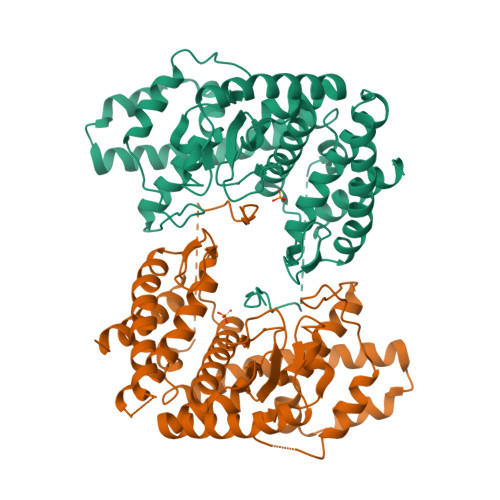
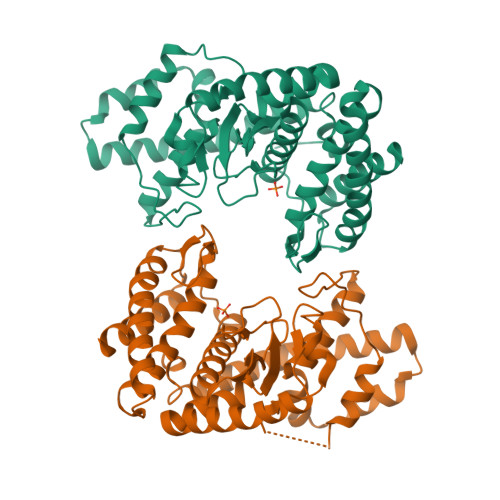
Fused radical SAM and alpha KG-HExxH domain proteins contain a distinct structural fold and catalyse cyclophane formation and beta-hydroxylation.
Morishita, Y., Ma, S., De La Mora, E., Li, H., Chen, H., Ji, X., Usclat, A., Amara, P., Sugiyama, R., Tooh, Y.W., Gunawan, G., Perard, J., Nicolet, Y., Zhang, Q., Morinaka, B.I.(2024) Nat Chem 16: 1882-1893
- PubMed: 39294420
- DOI: https://doi.org/10.1038/s41557-024-01596-9
- Primary Citation of Related Structures:
8S5F - PubMed Abstract:
Two of nature's recurring binding motifs in metalloproteins are the CxxxCxxC motif in radical SAM enzymes and the 2-His-1-carboxylate motif found both in zincins and α-ketoglutarate and non-haem iron enzymes. Here we show the confluence of these two domains in a single post-translational modifying enzyme containing an N-terminal radical S-adenosylmethionine domain fused to a C-terminal 2-His-1-carboxylate (HExxH) domain. The radical SAM domain catalyses three-residue cyclophane formation and is the signature modification of triceptides, a class of ribosomally synthesized and post-translationally modified peptides. The HExxH domain is a defining feature of zinc metalloproteases. Yet the HExxH motif-containing domain studied here catalyses β-hydroxylation and is an α-ketoglutarate non-haem iron enzyme. We determined the crystal structure for this HExxH protein at 2.8 Å, unveiling a distinct structural fold, thus expanding the family of α-ketoglutarate non-haem iron enzymes with a class that we propose to name αKG-HExxH. αKG-HExxH proteins represent a unique family of ribosomally synthesized and post-translationally modified peptide modifying enzymes that can furnish opportunities for genome mining, synthetic biology and enzymology.
Organizational Affiliation:
Department of Pharmacy, National University of Singapore, Singapore, Singapore.