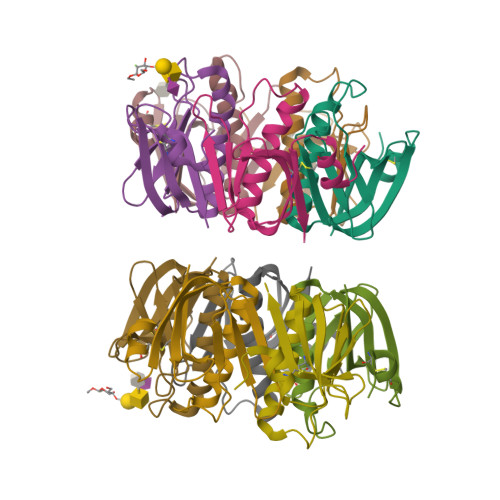
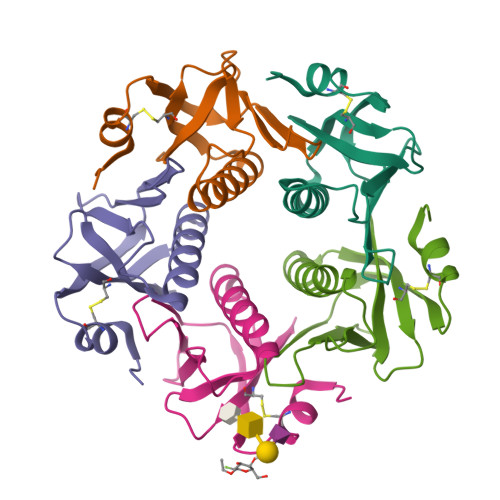
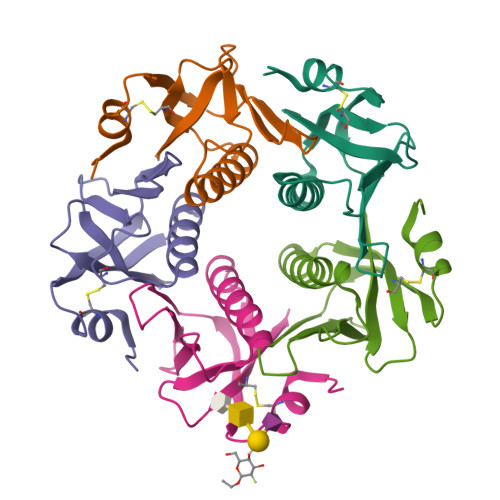
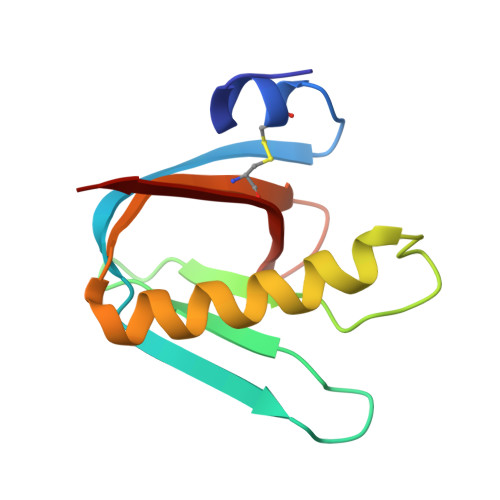
Probing the Origin of Affinity in the GM1-Cholera Toxin Complex through Site-Selective Editing with Fluorine.
Jordan, C., Hayashi, T., Lobbert, A., Fan, J., Teschers, C.S., Siebold, K., Aufiero, M., Pape, F., Campbell, E., Axer, A., Bussmann, K., Bergander, K., Kohnke, J., Gossert, A.D., Gilmour, R.(2024) ACS Cent Sci 10: 1481-1489
- PubMed: 39220706
- DOI: https://doi.org/10.1021/acscentsci.4c00622
- Primary Citation of Related Structures:
9EWF - PubMed Abstract:
Carbohydrates regulate an inimitable spectrum of biological functions, yet successfully leveraging this therapeutic avenue continues to be frustrated by low affinities with glycan-specific proteins. A conspicuous exception is the interaction of monosialotetrahexosylganglioside (GM1) with the carbohydrate-recognition domain of cholera toxin from Vibrio cholerae : this is one of the strongest protein-carbohydrate interactions known. To establish the importance of a long-discussed key hydrogen bond between C2 of the terminal galactose of GM1 and the B subunit pentamer of cholera toxin (CTB 5 ), the total synthesis of a selectively fluorinated GM1 epitope was conducted in 19 steps. This process of molecular editing (O δ- H → F δ- ) strategically deletes the hydrogen bond donor while retaining the localized partial charge of the substituent. Comparison of the binding affinity of F-GM1/CTB 5 with native GM1, the GM1 carbohydrate epitope, and meta -mononitrophenyl-α-galactoside (MNPG) revealed a trend that fully supports the importance of this key interaction. These NMR data suggest that F-GM1 binds in a closely similar conformation as native GM1. Crystallographic analyses of the complex also confirm that the OH → F bioisosteric exchange at C2 of the terminal galactose induces a ring conformation that eliminates key hydrogen bonds: these interactions are compensated for by inter- and intramolecular fluorine-specific interactions.
Organizational Affiliation:
Institute for Organic Chemistry, University of Münster, 48149 Münster, Germany.