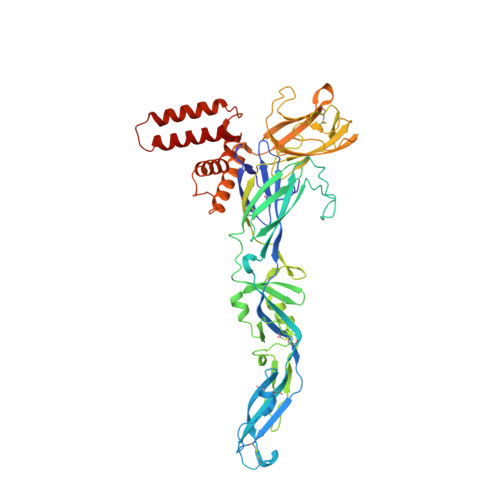
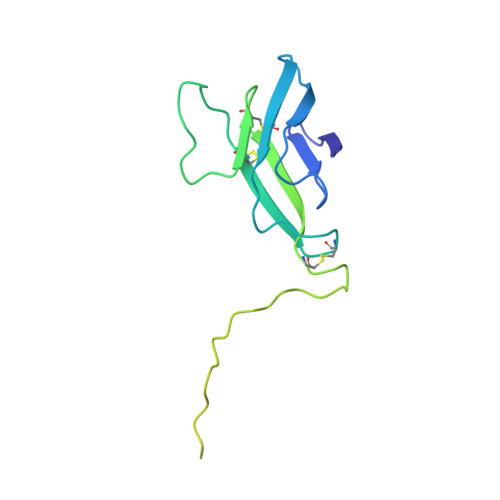
Flavivirus maturation leads to the formation of an occupied lipid pocket in the surface glycoproteins.
Renner, M., Dejnirattisai, W., Carrique, L., Martin, I.S., Karia, D., Ilca, S.L., Ho, S.F., Kotecha, A., Keown, J.R., Mongkolsapaya, J., Screaton, G.R., Grimes, J.M.(2021) Nat Commun 12: 1238-1238
- PubMed: 33623019
- DOI: https://doi.org/10.1038/s41467-021-21505-9
- Primary Citation of Related Structures:
6ZQI, 6ZQJ, 6ZQU, 6ZQV, 6ZQW - PubMed Abstract:
Flaviviruses such as Dengue (DENV) or Zika virus (ZIKV) assemble into an immature form within the endoplasmatic reticulum (ER), and are then processed by furin protease in the trans-Golgi. To better grasp maturation, we carry out cryo-EM reconstructions of immature Spondweni virus (SPOV), a human flavivirus of the same serogroup as ZIKV. By employing asymmetric localised reconstruction we push the resolution to 3.8 Å, enabling us to refine an atomic model which includes the crucial furin protease recognition site and a conserved Histidine pH-sensor. For direct comparison, we also solve structures of the mature forms of SPONV and DENV to 2.6 Å and 3.1 Å, respectively. We identify an ordered lipid that is present in only the mature forms of ZIKV, SPOV, and DENV and can bind as a consequence of rearranging amphipathic stem-helices of E during maturation. We propose a structural role for the pocket and suggest it stabilizes mature E.
Organizational Affiliation:
Division of Structural Biology, The Wellcome Centre for Human Genetics, University of Oxford, Oxford, UK.