Interpenetrating Cubes in the X-ray Crystallographic Structure of a Peptide Derived from Medin 19-36 .
Howitz, W.J., Wierzbicki, M., Cabanela, R.W., Saliba, C., Motavalli, A., Tran, N., Nowick, J.S.(2020) J Am Chem Soc 142: 15870-15875
- PubMed: 32816461
- DOI: https://doi.org/10.1021/jacs.0c06143
- Primary Citation of Related Structures:
7JRH - PubMed Abstract:
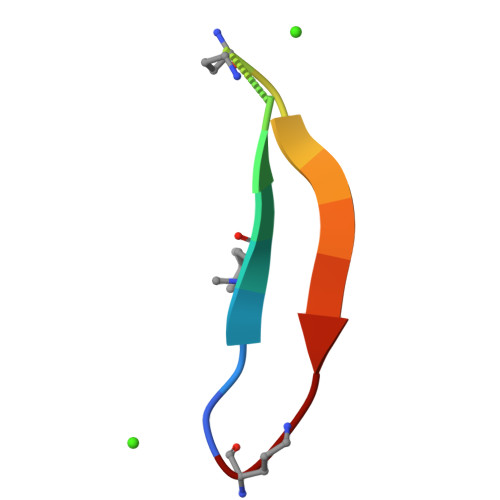
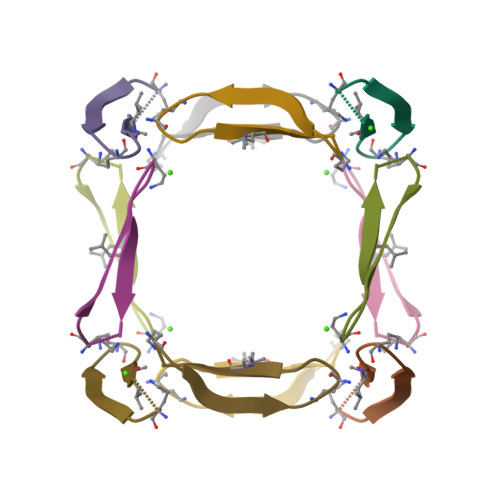
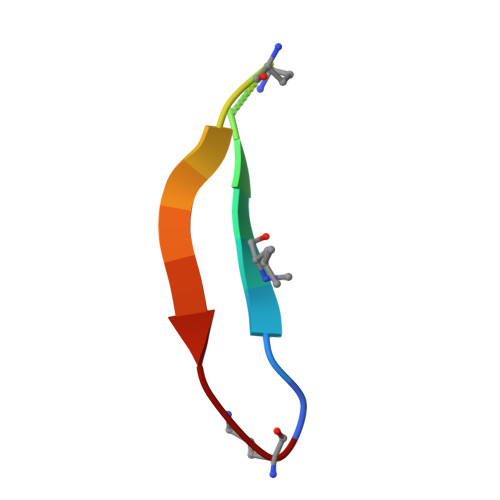
Amyloidogenic peptides and proteins are rich sources of supramolecular assemblies. Sequences derived from well-known amyloids, including Aβ, human islet amyloid polypeptide, and tau have been found to assemble as fibrils, nanosheets, ribbons, and nanotubes. The supramolecular assembly of medin, a 50-amino acid peptide that forms fibrillary deposits in aging human vasculature, has not been heavily investigated. In this work, we present an X-ray crystallographic structure of a cyclic β-sheet peptide derived from the 19-36 region of medin that assembles to form interpenetrating cubes. The edge of each cube is composed of a single peptide, and each vertex is occupied by a divalent metal ion. This structure may be considered a metal-organic framework (MOF) containing a large peptide ligand. This work demonstrates that peptides containing Glu or Asp that are preorganized to adopt β-hairpin structures can serve as ligands and assemble with metal ions to form MOFs.
Organizational Affiliation:
Department of Chemistry and Department of Pharmaceutical Sciences, University of California, Irvine, Irvine, California 92697-2025, United States.