Copper inactivates DcsB by oxidation of the Cys86 to cysteine sulfinic aicd
Oda, K., Komaguchi, K., Matoba, Y.To be published.
Experimental Data Snapshot
wwPDB Validation 3D Report Full Report
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| N(omega)-hydroxy-L-arginine amidinohydrolase | 281 | Streptomyces lavendulae | Mutation(s): 0 Gene Names: dcsB EC: 3.5.3.25 | 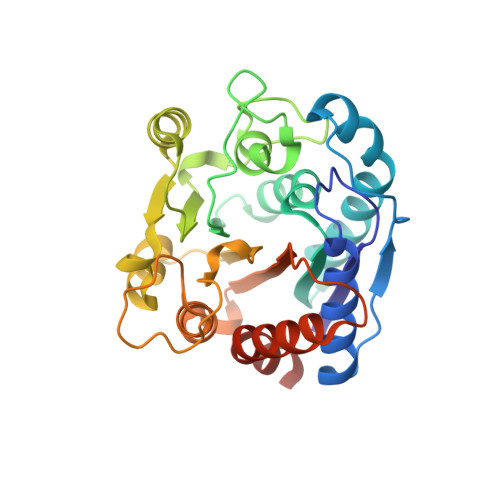 | |
UniProt | |||||
Find proteins for D2Z025 (Streptomyces lavendulae) Explore D2Z025 Go to UniProtKB: D2Z025 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | D2Z025 | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Ligands 4 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| CU1 Query on CU1 | G [auth B], H [auth B] | COPPER (I) ION Cu VMQMZMRVKUZKQL-UHFFFAOYSA-N |  | ||
| CU (Subject of Investigation/LOI) Query on CU | F [auth B] | COPPER (II) ION Cu JPVYNHNXODAKFH-UHFFFAOYSA-N |  | ||
| MN (Subject of Investigation/LOI) Query on MN | C [auth A], D [auth A] | MANGANESE (II) ION Mn WAEMQWOKJMHJLA-UHFFFAOYSA-N |  | ||
| MG (Subject of Investigation/LOI) Query on MG | E [auth A] | MAGNESIUM ION Mg JLVVSXFLKOJNIY-UHFFFAOYSA-N |  | ||
| Length ( Å ) | Angle ( ˚ ) |
|---|---|
| a = 46.502 | α = 83.57 |
| b = 46.981 | β = 84.69 |
| c = 59.559 | γ = 70.09 |
| Software Name | Purpose |
|---|---|
| PHENIX | refinement |
| Aimless | data scaling |
| XDS | data reduction |
| PHENIX | phasing |
| Funding Organization | Location | Grant Number |
|---|---|---|
| Not funded | -- |