New Features
Structural variation in clusters from PDBFlex
06/20
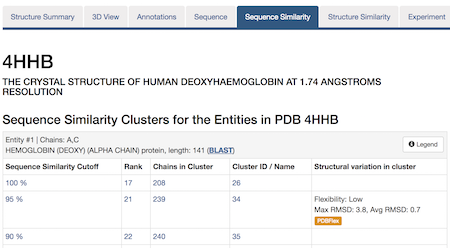
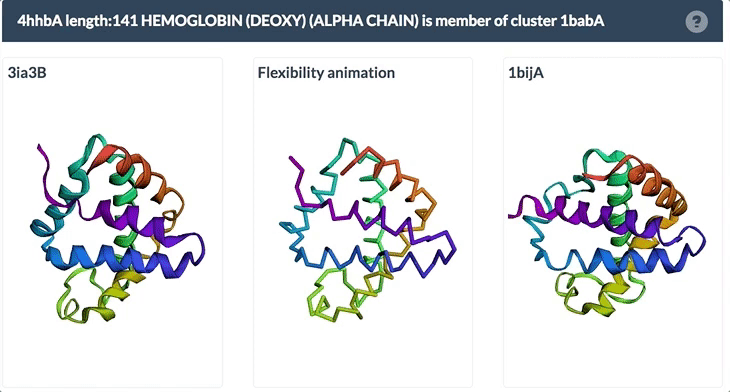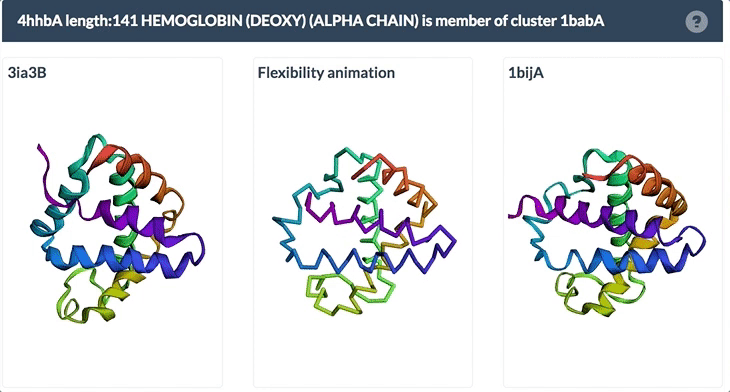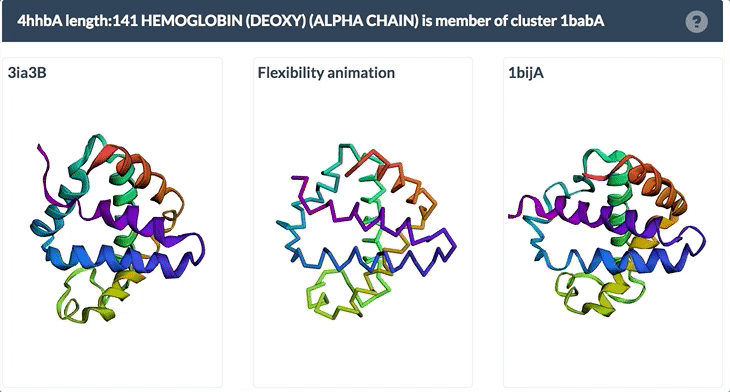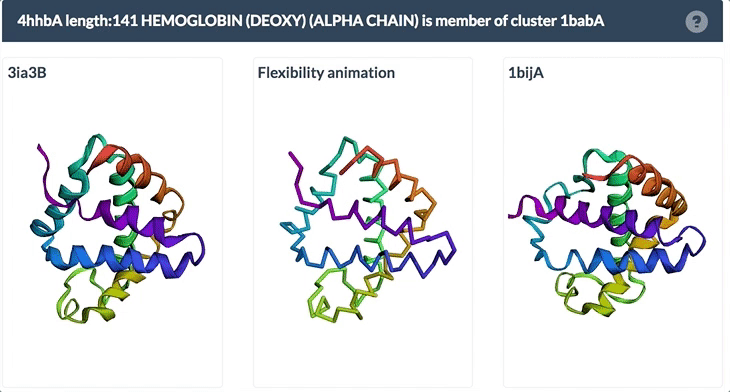
The same protein may display conformational flexibility in different experimental structures. Information regarding structural variation in similar sequences is now available on the Sequence Similarity tab of the Structure Summary Page through the integration of data from the PDBFlex database (pdbflex.org).
The PDBFlex database explores the intrinsic flexibility of protein structures by analyzing structural variations of the same protein across the archive. This allows for the easy identification of regions and types of structural flexibility present in a protein of interest. Structures of protein chains with identical sequences (sequence identity > 95%) are aligned, superimposed and clustered.
The example below shows the PDBFlex link from the Sequence Similarity tab for entry 4hhb and a view from the PDBFlex page for cluster 1babA of which the α-chain of entry 4hhb is a member. Here, the two structures within the cluster that have the highest Cα RMSD are displayed as well as a transformation between the two that helps to visualize the maximal flexibility in the cluster.
The identification of similar sequences in this report is based on the clustering used by RCSB PDB.