New Features
Mol* Viewer: modern web app for 3D visualization and analysis of large biomolecular structures
07/06 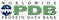

An article describing updates and improvements to Mol* is highlighted on the cover of the 2021 Nucleic Acids Research Web Server Issue. As the primary 3D structure viewer used by PDBe and RCSB PDB, it enables 3D exploration of macromolecular coordinate and experimental data directly within the browser window. The project is an open collaboration started by PDBe, RCSB PDB, CEITEC, and welcomes new contributors.
Two outstanding students, Áron Samuel Kovács and Sukolsak Sakshuwong, contributed new functionality to Mol*. Thanks to their work, Mol* now has greatly improved 3D rendering capabilities and can also export molecular scenes as 3D object files for use in external rendering programs. These features can currently be previewed on molstar.org before they are made available at PDBe and RCSB PDB.
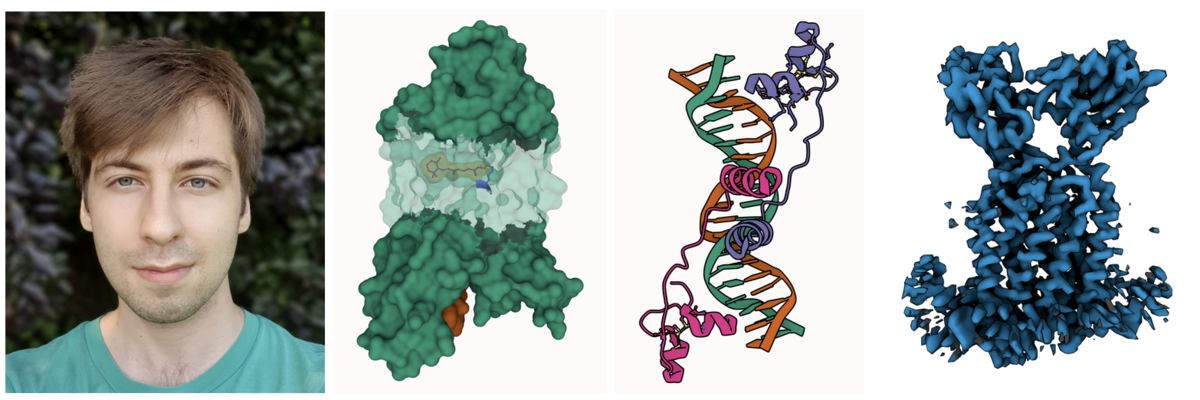 Photo of Áron; Partial transparency revealing retinal and surrounding channels in 3PQR; Stylized depiction of PDB ID 1D66 with crisp outlines; and Cryo-EM density of EMD-12604 with shadowing of occluded crevices.
Photo of Áron; Partial transparency revealing retinal and surrounding channels in 3PQR; Stylized depiction of PDB ID 1D66 with crisp outlines; and Cryo-EM density of EMD-12604 with shadowing of occluded crevices.Áron Samuel Kovács just finished his Master thesis in computer graphics at Masaryk University in the group of Barbora Kozlíková. He greatly improved the 3D rendering capabilities of Mol*, including artifacts-free transparency, improved darkening of crevices for better depth perception and much cleaner outlines.
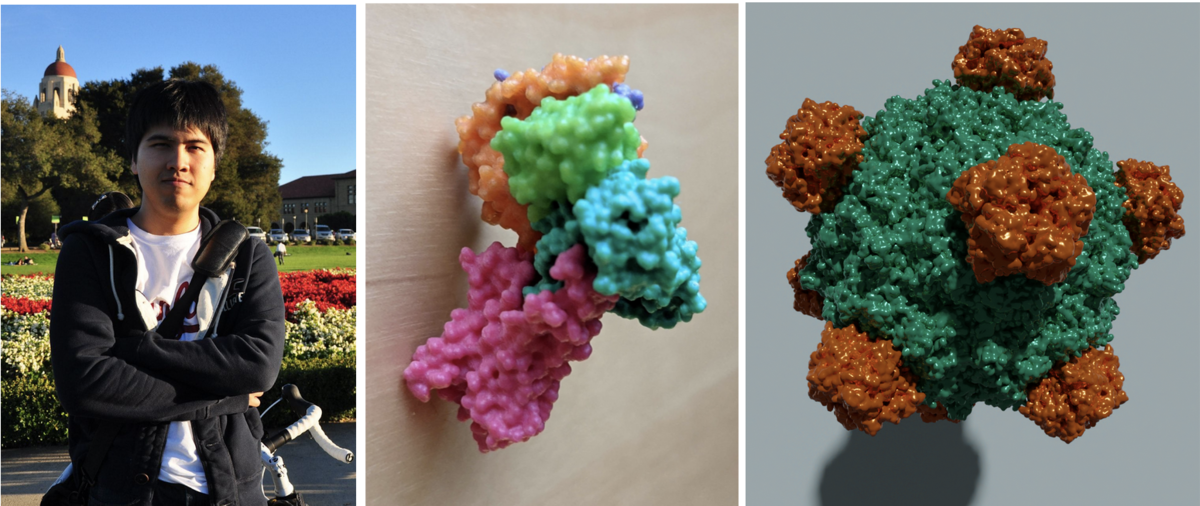 Photo of Sukolsak. 3D printed model of PDB ID 3SN6. High quality image of PDB ID 1RB8 rendered with the free and open source tool Blender.
Photo of Sukolsak. 3D printed model of PDB ID 3SN6. High quality image of PDB ID 1RB8 rendered with the free and open source tool Blender.Sukolsak Sakshuwong just finished his PhD in Management Science and Engineering at Stanford University in the group of Ashish Goel. He added geometry exporters to Mol* which allows users to extract 3D molecular scenes created in Mol* for use in 3D printing and other 3D graphic design. These scenes can be exported as glTF, an industry standard file format, as well as STL and Wavefront (.obj) formats.
The Mol* toolkit is available open access on GitHub, allowing community contributions.












