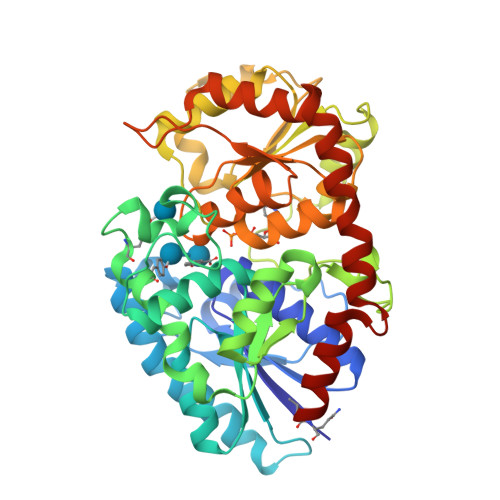
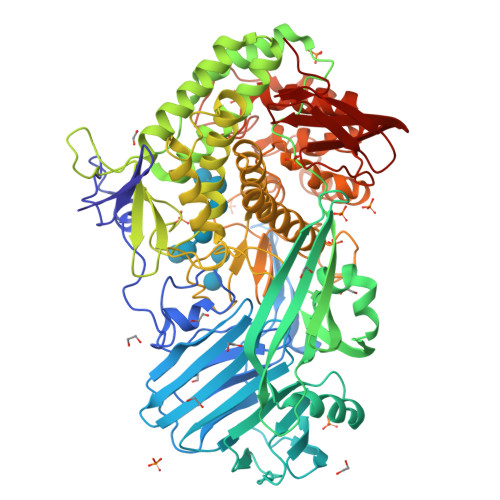
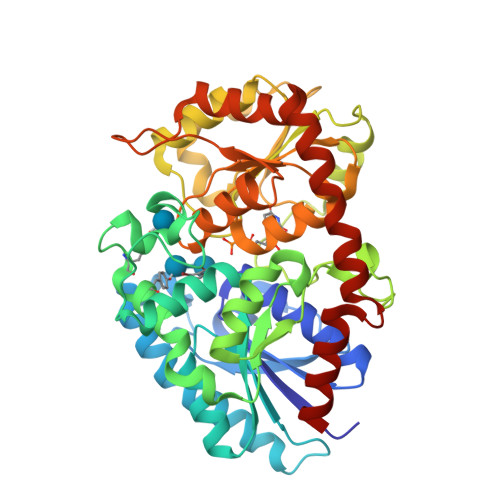
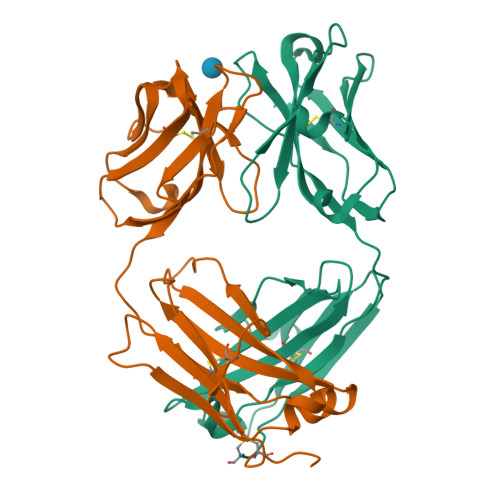
Structure Determination MethodologyScientific Name of Source OrganismMore... Refinement Resolution (Å)Enzyme Classification NameMembrane Protein Annotation | Boraston, A.B., Notenboom, V., Warren, R.A.J., Kilburn, D.G., Rose, D.R., Davies, G.J. (2003) J Mol Biology 327: 659 | Released | 2003-03-13 | | Method | X-RAY DIFFRACTION 1 Å | | Organisms | | | Macromolecule | | | Unique Ligands | ACY, CA | | Unique branched monosaccharides | BGC |
Meagher, J.L., Winter, H.C., Ezell, P., Goldstein, I.J., Stuckey, J.A. (2005) Glycobiology 15: 1033 | Released | 2005-06-16 | | Method | X-RAY DIFFRACTION 2.8 Å | | Organisms | | | Macromolecule | | | Unique Ligands | CD, SO4 | | Unique branched monosaccharides | BGC |
Money, V.A., Smith, N.L., Scaffidi, A., Stick, R.V., Gilbert, H.J., Davies, G.J. (2006) Angew Chem Int Ed Engl 45: 5136 | Released | 2006-07-12 | | Method | X-RAY DIFFRACTION 1.4 Å | | Organisms | | | Macromolecule | | | Unique Ligands | ZZ1 | | Unique branched monosaccharides | BGC |
Vasur, J., Kawai, R., Andersson, E., Igarashi, K., Sandgren, M., Samejima, M., Stahlberg, J. (2009) FEBS J 276: 3858 | Released | 2009-07-21 | | Method | X-RAY DIFFRACTION 1.1 Å | | Organisms | | | Macromolecule | | | Unique Ligands | LGC, NAG | | Unique branched monosaccharides | BGC |
Chuenchor, W., Pengthaisong, S., Robinson, R.C., Yuvaniyama, J., Svasti, J., Ketudat Cairns, J.R. (2011) J Struct Biol 173: 169-179 | Released | 2010-06-02 | | Method | X-RAY DIFFRACTION 2.8 Å | | Organisms | | | Macromolecule | | | Unique Ligands | MES, SO4, ZN | | Unique branched monosaccharides | BGC |
Chuenchor, W., Ketudat Cairns, J.R., Pengthaisong, S., Robinson, R.C., Yuvaniyama, J., Chen, C.-J. (2011) J Struct Biol 173: 169-179 | Released | 2009-11-03 | | Method | X-RAY DIFFRACTION 1.37 Å | | Organisms | | | Macromolecule | | | Unique Ligands | MES, SO4, ZN | | Unique branched monosaccharides | BGC |
Feinberg, H., Taylor, M.E., Razi, N., McBride, R., Knirel, Y.A., Graham, S.A., Drickamer, K., Weis, W.I. (2011) J Mol Biology 405: 1027-1039 | Released | 2010-12-08 | | Method | X-RAY DIFFRACTION 1.6051 Å | | Organisms | | | Macromolecule | | | Unique Ligands | BGC, CA | | Unique branched monosaccharides | BGC |
Kanagawa, M., Yamaguchi, Y. (2014) J Biological Chem 289: 16954-16965 | Released | 2013-09-25 | | Method | X-RAY DIFFRACTION 2 Å | | Organisms | | | Macromolecule | | | Unique Ligands | CL | | Unique branched monosaccharides | BGC |
Kataoka, M., Ishikawa, K. To be published | Released | 2015-09-16 | | Method | X-RAY DIFFRACTION 1.45 Å | | Organisms | | | Macromolecule | | | Unique Ligands | BGC, CA, GOL, NHE | | Unique branched monosaccharides | BGC |
Munshi, P., Cuneo, M.J. (2013) BMC Struct Biol 13: 18-18 | Released | 2013-10-09 | | Method | X-RAY DIFFRACTION 2.05 Å | | Organisms | | | Macromolecule | | | Unique Ligands | CA | | Unique branched monosaccharides | BGC |
Patskovsky, Y., Toro, R., Bhosle, R., Al Obaidi, N., Chamala, S., Attonito, J.D., Scott Glenn, A., Chowdhury, S., Lafleur, J., Siedel, R.D., Hillerich, B., Love, J., Whalen, K.L., Gerlt, J.A., Almo, S.C., Enzyme Function Initiative (EFI) To be published | Released | 2014-08-27 | | Method | X-RAY DIFFRACTION 2 Å | | Organisms | | | Macromolecule | | | Unique branched monosaccharides | BGC |
Qin, Z., Yan, Q., Lei, J., Yang, S., Jiang, Z. (2015) Acta Crystallogr D Biol Crystallogr 71: 1714-1724 | Released | 2015-08-12 | | Method | X-RAY DIFFRACTION 2.27 Å | | Organisms | | | Macromolecule | | | Unique branched monosaccharides | BGC |
Pluvinage, B., Boraston, A.B. (2017) Structure 25: 1348-1359.e3 | Released | 2017-06-28 | | Method | X-RAY DIFFRACTION 1.8 Å | | Organisms | | | Macromolecule | | | Unique Ligands | EDO, PO4 | | Unique branched monosaccharides | BGC, GLC |
George Thompson, A.M., Iancu, C.V., Dean, J.V., Choe, J. (2017) Sci Rep 7: 46629-46629 | Released | 2017-05-03 | | Method | X-RAY DIFFRACTION 2.001 Å | | Organisms | | | Macromolecule | | | Unique Ligands | BGC, GLN, SAL, UDP | | Unique branched monosaccharides | BGC |
Pluvinage, B., Boraston, A.B. (2017) Structure | Released | 2017-06-28 | | Method | X-RAY DIFFRACTION 1.65 Å | | Organisms | | | Macromolecule | | | Unique Ligands | EDO, PO4 | | Unique branched monosaccharides | BGC |
Pluvinage, B., Boraston, A.B. (2017) Structure | Released | 2017-06-28 | | Method | X-RAY DIFFRACTION 1.7 Å | | Organisms | | | Macromolecule | | | Unique Ligands | EDO, PO4 | | Unique branched monosaccharides | BGC, GLC |
Pluvinage, B., Boraston, A.B. (2017) Structure | Released | 2017-06-28 | | Method | X-RAY DIFFRACTION 2.15 Å | | Organisms | | | Macromolecule | | | Unique Ligands | EDO, PO4 | | Unique branched monosaccharides | BGC |
|