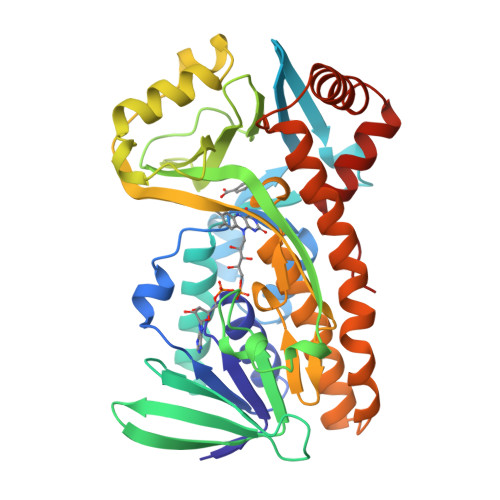
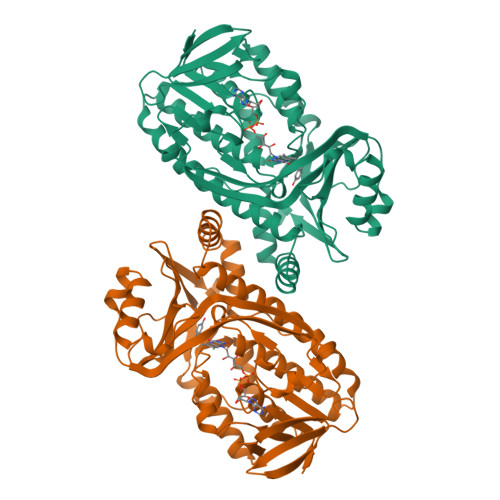
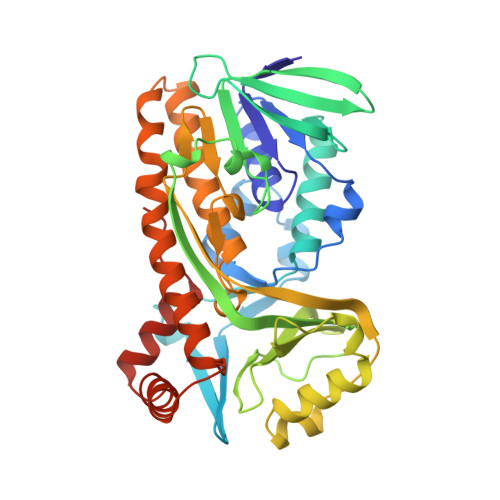
Interdomain binding of NADPH in p-hydroxybenzoate hydroxylase as suggested by kinetic, crystallographic and modeling studies of histidine 162 and arginine 269 variants.
Eppink, M.H., Schreuder, H.A., van Berkel, W.J.(1998) J Biological Chem 273: 21031-21039
- PubMed: 9694855
- DOI: https://doi.org/10.1074/jbc.273.33.21031
- Primary Citation of Related Structures:
1BGJ, 1BGN - PubMed Abstract:
The conserved residues His-162 and Arg-269 of the flavoprotein p-hydroxybenzoate hydroxylase (EC 1.14.13.2) are located at the entrance of the interdomain cleft that leads toward the active site. To study their putative role in NADPH binding, His-162 and Arg-269 were selectively changed by site-specific mutagenesis. The catalytic properties of H162R, H162Y, and R269K were similar to the wild-type enzyme. However, less conservative His-162 and Arg-269 replacements strongly impaired NADPH binding without affecting the conformation of the flavin ring and the efficiency of substrate hydroxylation. The crystal structures of H162R and R269T in complex with 4-hydroxybenzoate were solved at 3.0 and 2.0 A resolution, respectively. Both structures are virtually indistinguishable from the wild-type enzyme-substrate complex except for the substituted side chains. In contrast to wild-type p-hydroxybenzoate hydroxylase, H162R is not inactivated by diethyl pyrocarbonate. NADPH protects wild-type p-hydroxybenzoate hydroxylase from diethylpyrocarbonate inactivation, suggesting that His-162 is involved in NADPH binding. Based on these results and GRID calculations we propose that the side chains of His-162 and Arg-269 interact with the pyrophosphate moiety of NADPH. An interdomain binding mode for NADPH is proposed which takes a novel sequence motif (Eppink, M. H. M., Schreuder, H. A., and van Berkel, W. J. H. (1997) Protein Sci. 6, 2454-2458) into account.
Organizational Affiliation:
Department of Biomolecular Sciences, Laboratory of Biochemistry, Wageningen Agricultural University, Dreijenlaan 3, 6703 HA Wageningen, The Netherlands.