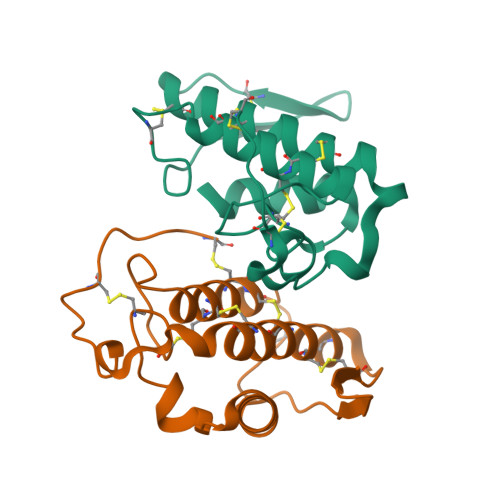
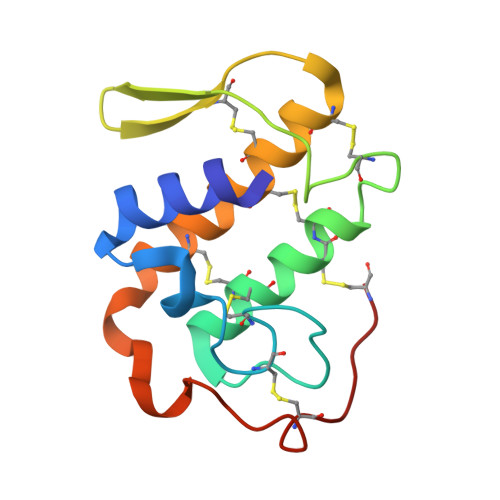
Three-dimensional structure of a presynaptic neurotoxic phospholipase A2 from Daboia russelli pulchella at 2.4 A resolution.
Chandra, V., Kaur, P., Srinivasan, A., Singh, T.P.(2000) J Mol Biology 296: 1117-1126
- PubMed: 10686108
- DOI: https://doi.org/10.1006/jmbi.2000.3537
- Primary Citation of Related Structures:
1CL5 - PubMed Abstract:
The phospholipase A(2 )from Daboia russelli pulchella (DPLA(2)) is the only known member of subclass II of group IIA. The three-dimensional structure of this presynaptic neurotoxic DPLA(2) enzyme has been determined at 2.4 A resolution. The structure was determined by the molecular replacement method using the model Crotalus atrox, and refined using X-PLOR to a final R-factor of 18.8 % for all data in the resolution range 20.0 A-2.4 A. The final refined model comprises 1888 atoms from two crystallographically independent protein molecules and 160 water oxygen atoms. The overall folding of DPLA(2), with three long helices and two short antiparallel beta-strands is grossly similar to those observed for other PLA(2)s. In the present structure, the calcium binding site is empty but the conformation of the calcium binding loop is similar to those observed in the calcium bound states. Two spatially adjacent regions of residues 55-61 (a typical beta-turn I) and 83-94 (a well defined loop) are remarkably different in conformation, electrostatic characteristics and inter-segmental interactions from those found in non-neurotoxic PLA(2)s. Yet another striking structural feature in DPLA(2 )pertains to the stretch of residues 53-77, which has a series of positively charged residues protruding outwardly. The above segment is presumed to be involved in the anticoagulant activity. A unique hydrophobic patch including residues Leu17, Ala18, Ile19, Pro20, Phe106 and Leu110 is found on the surface together with an equally emphatic region of -OH groups containing residues such as Ser21, Tyr22, Ser23, Ser24, Tyr25 and Tyr28. The interactions between two molecules of DPLA(2) in the asymmetric unit are remarkably different from those observed in the standard dimers and trimers of PLA(2)s, leaving the enzyme's active site fully exposed for enzyme-substrate reactions, it makes this structure one of the most favourable examples for structure-based drug design through soaking experiments.
Organizational Affiliation:
Department of Biophysics, All India Institute of Medical Sciences, New Delhi, 110 029, India.