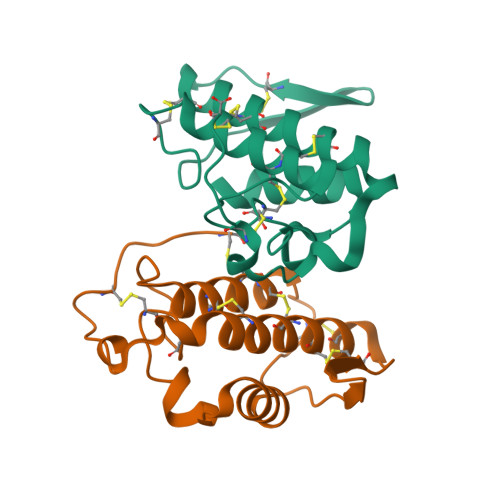
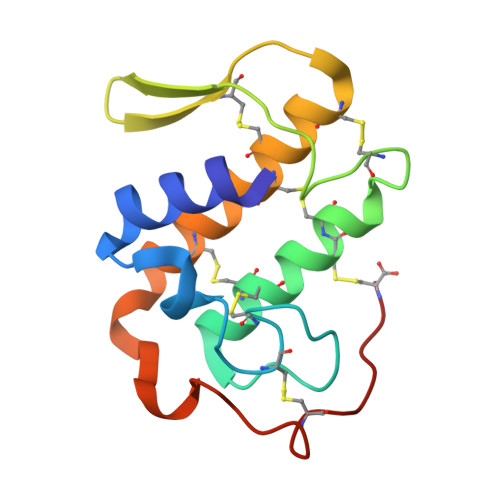
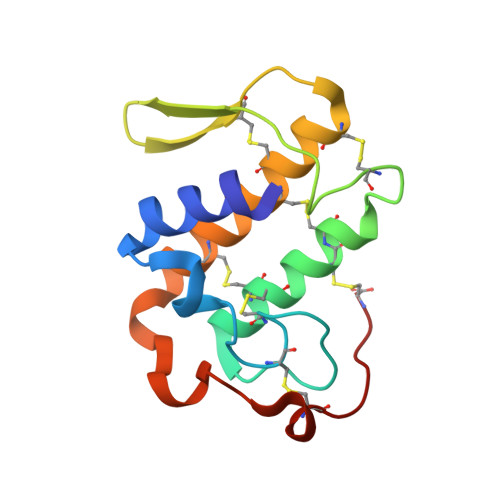
Regulation of catalytic function by molecular association: structure of phospholipase A2 from Daboia russelli pulchella (DPLA2) at 1.9 A resolution.
Chandra, V., Kaur, P., Jasti, J., Betzel, C., Singh, T.P.(2001) Acta Crystallogr D Biol Crystallogr 57: 1793-1798
- PubMed: 11717491
- DOI: https://doi.org/10.1107/s0907444901014524
- Primary Citation of Related Structures:
1FB2 - PubMed Abstract:
The crystal structure of phospholipase A(2) from the venom of Daboia russelli pulchella has been refined to an R factor of 0.216 using 17,922 reflections to 1.9 A resolution. The structure contains two crystallographically independent molecules in the asymmetric unit. The overall conformations of the two molecules are essentially the same except for three regions, namely the calcium-binding loop including Trp31, the beta-wing and the C-terminal residues 119-131. Although these differences have apparently been caused by molecular packing, they seem to have functional relevance. Particularly noteworthy is the conformation of Trp31, which is favourable for substrate binding in one molecule as it is aligned with one of the side walls of the hydrophobic channel, whereas in the other molecule it is located at the mouth of the channel, thereby blocking the entry of substrates leading to loss of activity. This feature is unique to the present structure and does not occur in the dimers and trimers of other PLA(2)s.
Organizational Affiliation:
Department of Biophysics, All India Institute of Medical Sciences, New Delhi 110029, India.