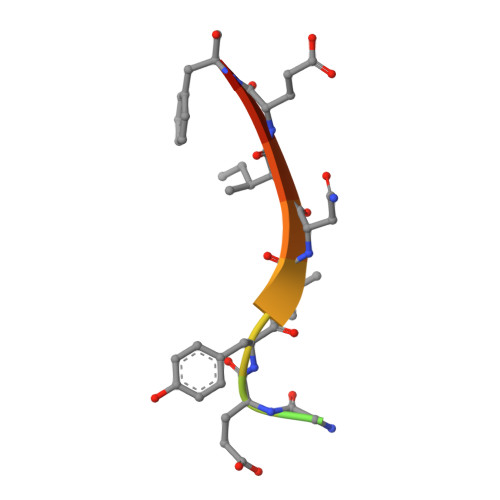
Structure and autoregulation of the insulin-like growth factor 1 receptor kinase.
Favelyukis, S., Till, J.H., Hubbard, S.R., Miller, W.T.(2001) Nat Struct Biol 8: 1058-1063
- PubMed: 11694888
- DOI: https://doi.org/10.1038/nsb721
- Primary Citation of Related Structures:
1K3A - PubMed Abstract:
The insulin-like growth factor 1 (IGF1) receptor is closely related to the insulin receptor. However, the unique biological functions of IGF1 receptor make it a target for therapeutic intervention in human cancer. Using its isolated tyrosine kinase domain, we show that the IGF1 receptor is regulated by intermolecular autophosphorylation at three sites within the kinase activation loop. Steady-state kinetic analyses of the isolated phosphorylated forms of the IGF1 receptor kinase (IGF1RK) reveal that each autophosphorylation event increases enzyme turnover number and decreases Km for ATP and peptide. We have determined the 2.1 A-resolution crystal structure of the tris-phosphorylated form of IGF1RK in complex with an ATP analog and a specific peptide substrate. The structure of IGF1RK reveals how the enzyme recognizes peptides containing hydrophobic residues at the P+1 and P+3 positions and how autophosphorylation stabilizes the activation loop in a conformation that facilitates catalysis. Although the nucleotide binding cleft is conserved between IGF1RK and the insulin receptor kinase, sequence differences in the nearby interlobe linker could potentially be exploited for anticancer drug design.
Organizational Affiliation:
Department of Physiology and Biophysics, School of Medicine State University of New York at Stony Brook, Stony Brook, New York 11794, USA.